-
Notifications
You must be signed in to change notification settings - Fork 7
Transcript Assembly Merge
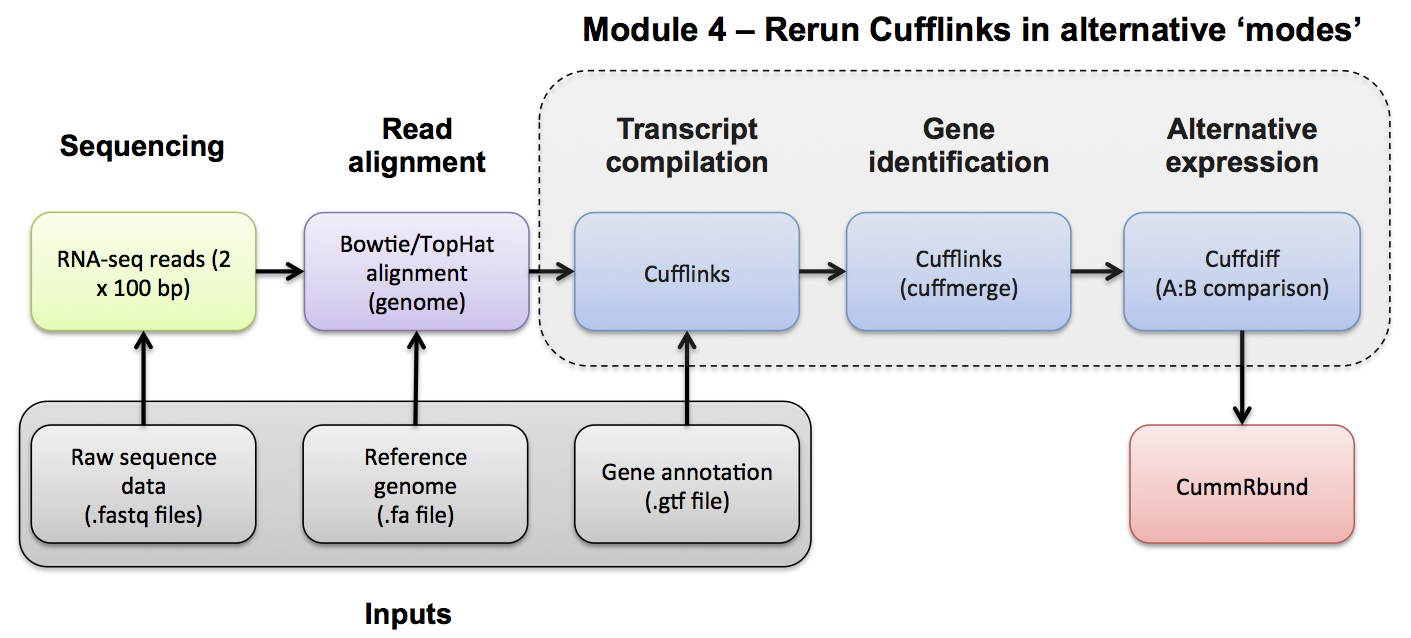
#4-iii. CUFFMERGE Use Cuffmerge to merge predicted transcripts from all libraries into a unified transcriptome. Refer to the Cufflinks manual for a more detailed explanation:
http://cole-trapnell-lab.github.io/cufflinks/cuffmerge/index.html
Cuffmerge basic usage
cuffmerge [options]* <assembly_GTF_list.txt>
Options specified below:
- "assembly_GTF_list.txt" is a text file "manifest" with a list (one per line) of GTF files that you would like to merge together into a single GTF file.
- '-p 8' tells cuffmerge to use eight CPUs
- '-o' tells cuffmerge to write output to a particular directory
- '-g' tells cuffmerge where to find reference gene annotations. It will use these annotations to gracefully merge novel isoforms (for de novo runs) and known isoforms and maximize overall assembly quality.
- '-s' tells cuffmerge where to find the reference genome files
Merge all 4 cufflinks results so that they will have the same set of transcripts for comparison purposes:
For reference guided mode:
cd $RNA_HOME/expression/tophat_cufflinks/ref_guided/
ls -1 *Rep*_ERCC*/transcripts.gtf > assembly_GTF_list.txt
cat assembly_GTF_list.txt
cuffmerge -p 8 -o merged -g $RNA_HOME/refs/hg19/genes/genes_chr22_ERCC92.gtf -s $RNA_HOME/refs/hg19/bwt/chr22_ERCC92/ assembly_GTF_list.txt
For de novo mode:
cd $RNA_HOME/expression/tophat_cufflinks/de_novo/
ls -1 *Rep*_ERCC*/transcripts.gtf > assembly_GTF_list.txt
cat assembly_GTF_list.txt
cuffmerge -p 8 -o merged -g $RNA_HOME/refs/hg19/genes/genes_chr22_ERCC92.gtf -s $RNA_HOME/refs/hg19/bwt/chr22_ERCC92/ assembly_GTF_list.txt
| Previous Section | This Section | Next Section | |:------------------------------------------------:|:------------------------------------------:|:----------------------------------------------------:| | De novo | Merging | Differential Splicing |
##Note: The current version of this tutorial is now at www.rnaseq.wiki
Table of Contents
Module 0: Authors | Citation | Syntax | Intro to AWS | Log into AWS | Unix | Environment | Resources
Module 1: Installation | Reference Genomes | Annotations | Indexing | Data | Data QC
Module 2: Adapter Trim | Alignment | IGV | Alignment Visualization | Alignment QC
Module 3: Expression | Differential Expression | DE Visualization
Module 4: Ref Guided | De novo | Merging | Differential Splicing | Splicing Visualization
Module 5: Kallisto
Appendix: Abbreviations | Lectures | Practical Exercise Solutions | Integrated Assignment | Proposed Improvements | AWS Setup