-
Notifications
You must be signed in to change notification settings - Fork 3
Mechanism of Action: Possible PfATP4 Activity Deduced from Parasite Ion Regulation Assays
Plasmodium falciparum is an intraerythrocytic (situated or occurring within the red blood cells) malaria parasite. On entering a red blood cell, the environment experienced by the parasite changes from high Na+ concentration and low K+ concentration (blood plasma) to the low [Na+] and high [K+] media of the cytosol. Around 12 hours after invasion, the parasite establishes itself in the host and develops 'new permeability pathways' for the influx of Na+ and efflux of K+, which in turn provide the energy required transport nutrients across it's membrane. The new permeability routes lead to a change in erythroyte cytosol environment so that the ratio of Na+/K+ is again reversed to mirror that of blood plasma. Despite the change surrounding the parasite, it manages to maintain a low cytosolic [Na+] concentration. The mechanism of Na+ homeostasis within the Plasmodium falciparum parasite has been investigated by Prof. Kiaran Kirk's group at the Australian National University. Investigations by the Kirk lab have shown that the cation ATPase PfATP4 is responsible for maintaining low Na+ concentration in healthy parasites.
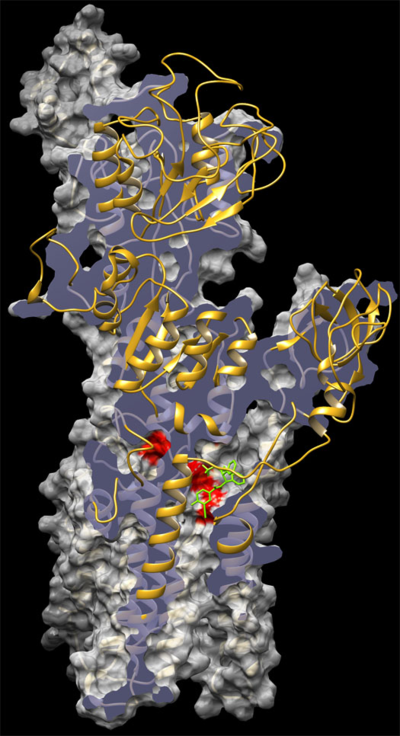
P. falciparum P-type cation-transporter ATPase 4, or PfATP4 is an integral parasite plasma membrane protein that was originally thought to be a Ca2+ pump. Current evidence suggests that PfATP4 is involved in [Na+] regulation. Currently, the structure of PfATP4 is unknown and Kirk's assays (measuring parasite [Na+]) are indirect measurements of postulated inhibition of PfATP4. Joseph DeRisi built a model of PfATP4 with KAE960 'docked in the presumptive sodium channel'. This is an example of a homology model, as it has been built using known structures of similar ion channels.
Figure: Homology model of PfATP4 with docked compound by J. DeRisi and J. Horst
Millions of compounds have been explored as a potential antimalarials as part of a concerted global effort to find new medicines to combat emerging resistance to existing drugs. Whole cell assays are an essential high throughput screening method for the identification of active compounds. In 2010, whole cell screening revealed a potent class of compounds, the spiroindolones, which were able to inhibit the growth of blood stage malaria parasites in vitro.
Following a barrage of other in vitro assays, one of the spirondolones, KAE690, was progressed to Phase II human clinical trials, where it was shown to be effective against uncomplicated Plasmodium falciparum and P. vivax. Additionally, the drug was active in late stage gametocyte assays. The ability to inhibit gametocytes is highly prized in antimalarial candidates as gametocytes are the sexual form of the parasite transmitted back to the mosquito. Therefore, drugs that terminate gametocytes play an important role in breaking the cycle of transmission.
KAE690 was next explored in drug pressure experiments. Microbial pathogens, such as plasmodium, are well known to develop resistance to drugs under prolonged exposure to medicines, both in laboratory experiments and in affected populations. Lab experiments can simulate the resistance developed in patients and also provide insight into the mechansims of resistance, which in turn can help to identify the mechanism of action of the therapeutic. Increasing concentrations of KAE609 were applied to particular parasite strain over the course of a few months. The amount of drug required to inhibit parasetemia by 50% (IC50), was found to increase, indicating that the parasite had developed some resistance to the drug. Genetic analysis showed that the 'resistant' parasite had several mutations in a gene located within it's plasma membrane, PfATP4.
The following five compounds inherited compounds were evaluated in parasite ion regulation assays in the Kirk Laboratory in 2013, with a subsequent set of seven compounds evaluated in 2014; the hypothesis is that PfATP4 is a Na+ ATPase that exports Na+ and imports H+ (or equivalent) and that the effects of the compounds on Na+ concentration and pH are attributable to inhibition of this activity. Structures, potency, metabolism/solubility and raw PfATP4 assay data are here.
(Compounds that are inactive do not dissipate the plasma membrane Na+ gradient or increase the plasma membrane pH gradient, consistent with them not inhibiting PfATP4 at the concentration tested. The other compounds dissipated the plasma membrane Na+ gradient and increased the plasma membrane pH gradient at a concentration of 2 μM, consistent with them being PfATP4 inhibitors.)
There is a correlation: compounds inactive in these assays are not potent vs the parasite.
Three Series 4 compounds were evaluated against four resistant strains (Dd2 parent and three different PfATP4 mutants) (ELN entry and GHI 251) in the laboratories of Kiaran Kirk and David Fidock data here. The compounds were found to have reduced efficacy against the PfATP4 mutants when compared to the parent Dd2 strain. These results have more recently been complemented by a similar experiment from Chase Smith and the Broad Institute, also indicating a shared MoA.
Following the synthesis of the 18 compounds for the Frontrunners campaign in 2016, these and another 18 novel compounds were evaluated in the Ion Regulation assay. Data is posted here.
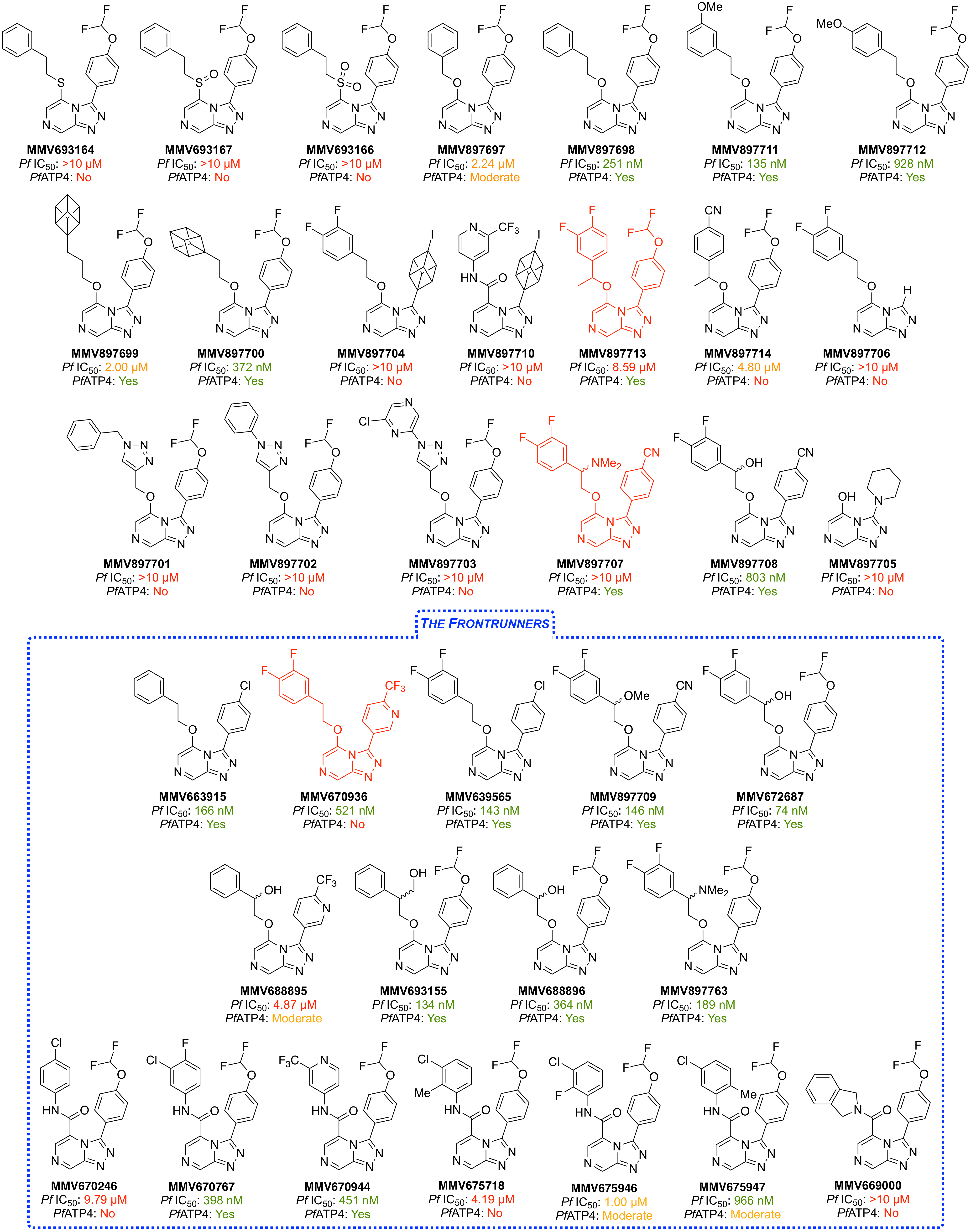
~92% of the 2016 data set shows correlation between potency and PfATP4 activity. However, the three compounds highlighted in red appear to not follow this trend, with them having activity in one assay but not the other.
PfATP4 is the apparent target of a number of different chemotypes.
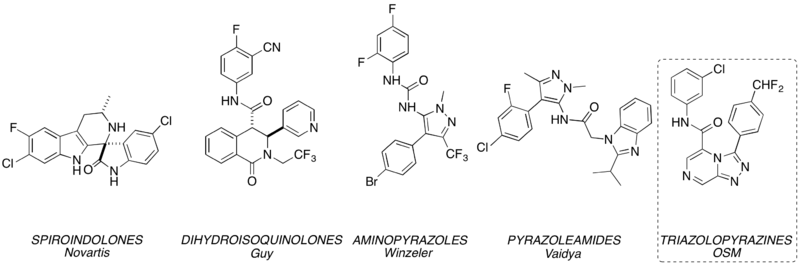
PfATP4 is implicated in the MoA of several other leading antimalarials in development: the spiroindolone KAE609, the pyrazoleamides, the dihydroisoquinolone (+)-SJ-733 and various aminopyrazoles. A striking diversity of other compounds (from the malaria box) behave in the same way (summarised here), leading to the question: is PfATP4 really the target? There is no physical proof of binding between any of these compounds and PfATP4, because the protein has not yet been generated pure or crystallised. There is a homology model. Cross-resistance has been seen between parasites grown to be resistant to compound X (where there are mutations that are associated with PfATP4) and then tested with compound Y, including for the Series 4 compounds, as above. A current project strand is to develop a pharmacophore model that throws light on which compounds will be active in Kiaran's assay, and how they might be binding to PfATP4 (in part to allow us to de-prioritise the development of any more compounds having this same target). An initial attempt at developing this model was unsuccessful (i.e. not predictive - see the figure, where the "P Model predictions" correlate poorly with what was found in the ion regulation assay) possibly because the model did not allow for overlapping binding sites or take into consideration compound chirality. Model generation now needs to be re-attempted, and OSM needs community expertise in this area to proceed.
Aims, Concerns and Current Interest in Series 4
Modification of Core Triazolopyrazine
Modification of Pyrazine Substitution Pattern
Modification of the Triazole Substitution
Pyrazine Side Chain Modifications - Ethers
Pyrazine Side Chain Modifications - Amides
Pyrazine Side Chain Modifications - Reversed Amides
Pyrazine Side Chain Modifications - Others
Biological Data Currently not Incorporated into the Main Wiki Sections
Mechanism of Action: Possible PfATP4 Activity Deduced from Parasite Ion Regulation Assays
Synthesis of the Ether-Linked Series
Synthesis of the Amide-Linked Series
Synthesis of the Reverse Amide- Linked Series
Synthesis of Benzylic Functionalised Ether-Linked Series
Alternative Routes to the Triazolopyrazine Core
Triazolopyrazine telesubstitution
Chirality/Stereogenic Centres in This Series
Other Sources of Compounds Relevant to this Series
Desirable Compounds Not Yet Synthesised
