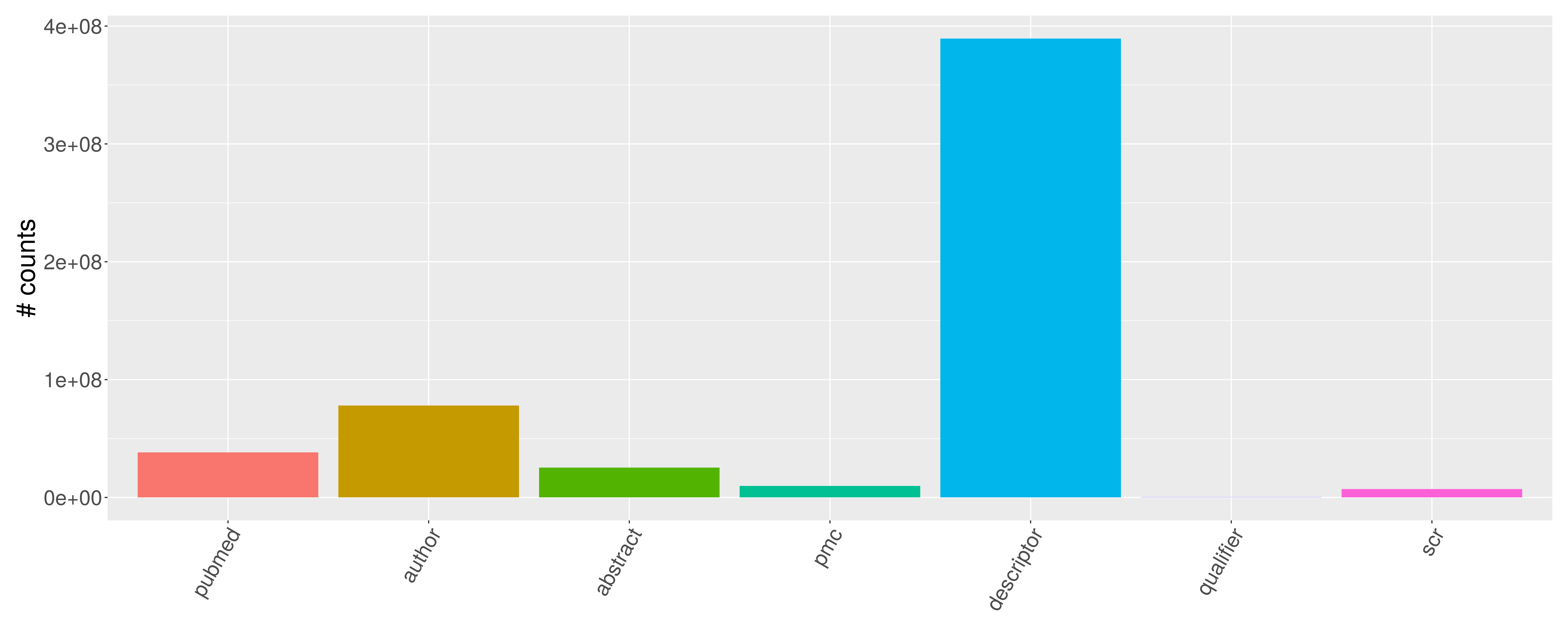
Workflow to construct to preprocess PubMed XML files and save sqlite, tibble, and data.table datasets.
- Bash: GNU bash, version 4.2.46(1)-release (x86_64-redhat-linux-gnu)
- Snakemake: 6.0.5
- Singularity: 3.5.3
- config.yaml:
- METADATA_VERSION: Update like v001 -> v002 -> ...and so on.
- BIOC_VERSION: Set next version of Bioconductor
The workflow consists of four snakemake workflows.
In local machine:
snakemake -s workflow/download.smk -j 4 --use-singularity
snakemake -s workflow/preprocess.smk -j 4 --use-singularity
snakemake -s workflow/metadata.smk -j 4 --use-singularity
snakemake -s workflow/plot.smk -j 4 --use-singularityIn parallel environment (GridEngine):
snakemake -s workflow/download.smk -j 4 --cluster "qsub -l nc=4 -p -50 -r yes -q node.q" --latency-wait 600 --use-singularity
snakemake -s workflow/preprocess.smk -j 4 --cluster "qsub -l nc=4 -p -50 -r yes -q node.q" --latency-wait 600 --use-singularity
snakemake -s workflow/metadata.smk -j 4 --cluster "qsub -l nc=4 -p -50 -r yes -q node.q" --latency-wait 600 --use-singularity
snakemake -s workflow/plot.smk -j 4 --cluster "qsub -l nc=4 -p -50 -r yes -q node.q" --latency-wait 600 --use-singularityIn parallel environment (Slurm):
snakemake -s workflow/download.smk -j 4 --cluster "sbatch -n 4 --nice=50 --requeue -p node03-06" --latency-wait 600 --use-singularity
snakemake -s workflow/preprocess.smk -j 4 --cluster "sbatch -n 4 --nice=50 --requeue -p node03-06" --latency-wait 600 --use-singularity
snakemake -s workflow/metadata.smk -j 4 --cluster "sbatch -n 4 --nice=50 --requeue -p node03-06" --latency-wait 600 --use-singularity
snakemake -s workflow/plot.smk -j 4 --cluster "sbatch -n 4 --nice=50 --requeue -p node03-06" --latency-wait 600 --use-singularityCopyright (c) 2021 Koki Tsuyuzaki and RIKEN Bioinformatics Research Unit Released under the Artistic License 2.0.
- Koki Tsuyuzaki
- Manabu Ishii
- Itoshi Nikaido