-
Notifications
You must be signed in to change notification settings - Fork 21
Retinotopy
Neuropythy has a particular focus on retinotopic mapping and properties of the visual cortex due to its author's interest in visual neuroscience. Most of the functionality relating to retinotopy and retinotopic maps is packaged in the neuropythy.vision package. This package contains many tools:
- Prediction of retinotopic maps, based on sulcal anatomy
- Bayesian inference of retinotopic maps, given a small amount of measured retinotopy
- Conversion of retinotopy fields between various forms (e.g., cartesian versus polar coordinates)
- Principled smoothing of retinotopic maps
(Note: this list is a work in progress and not complete.)
| Year | Reference | Description |
|---|---|---|
| 1909 | Inouye (1909)1 | In 1909 Tatsuji Inouye, a Japanese ophthalmologist, created the first retinotopic map by studying the relationship between brain injuries and visual field loss. His subjects were soldiers wounded in the Russo-Japanese war of 1904 and 1905. |
| 1918 | Holmes (1918)2 | In 1918, Gordon Holmes, a British neurologist, independently discovered retinotopic maps using the same method as Tatsuji Inouye. His subjects were British soldiers wounded in World War I. |
| 1994 | Engel et al., (1994) | Steven Engel and colleagues developed an expanding-ring and rotating-wedge stimulus used to produce high-quality retinotopic maps using non-invasive functional MRI. |
| 2008 | Dumoulin & Wandell, (2008) | Serge Dumoulin and Brian Wandell proposed a population receptive field (pRF) model of visual cortex that improves on previous travelling-wave methods of estimating retinotopic map parameters. |
| 2008 | Hinds et al., (2008) | Oliver Hinds and colleagues used FreeSurfer's cortical surface alignment to show that the boundary of V1 was well-predicted by anatomy. See Dale et al., (1999) and Fischl et al., (1999) for details on FreeSurfer's cortical surface alignment. |
| 2012 | Benson et al., (2012) | Noah Benson and colleagues built on the work of Hinds et al. by showing that the retinotopic organization of V1, in addition to its boundaries, was in register between subjects after anatomical alignment. |
| 2014 | Benson et al., (2014) | Noah Benson and colleagues constructed an anatomically-based retinotopic atlas of V1, V2, and V3--for a subject aligned to FreeSurfer's fsaverage atlas, the retinotopic maps in V1-V3 could be predicted using interpolation from this atlas. |
| 2015 | Wang et al., (2015) | Liang Wang and colleagues published an atlas of the boundaries of V1, V2, V3, hV4, V3a, V3b, LO1, LO2, VO1, VO2, and a number of other visual areas by aligning the cortical surfaces of 53 subjects using FreeSurfer. |
| 2018 | Benson & Winawer, (2018) | Noah Benson and Jonathan Winawer describe a Bayesian method that fits a model of retinotopic maps to the entire visual cortex rather than to individual voxels. |
1 Inouye T (1909) Die Sehstörungen bei Schussverletzungen der kortikalen Sehsphäre nach Beobachtungen an Verwundeten der letzten japanischen Kriege. (Engelmann, Leipzig)
2 Holmes G (1918) Disturbances of vision by cerebral lesions. Brit. J. Ophthal. 2:353-384
Prediction of retinotopic maps can be performed using either neuropythy's neuropythy.vision.predict_retinotopy function or the benson14_retinotopy command. The two methods use the same code but have different interfaces. Both apply the method described by Benson et al., (2014), which builds on previous work by Hinds et al., (2008). Both of these works used FreeSurfer's cortical-surface alignment method, in which subjects are aligned to an average atlas called the fsaverage. Once two subjects' cortical surfaces are aligned to this atlas, the functional organization of their cortices is also approximately aligned due to an approximate correspondence between anatomical structure and function. Accordingly, good estimates of a subejct's retinotopic maps may be made by aligning their cortices to the fsaverage atlas and interpolating the maps from the atlas onto the subject's cortical surfaces.
Both of neuropythy's methods for predicting retinotopy based on anatomy use an updated version of the atlas published by Benson et al., (2014). This newer atlas was constructed in a similar fashion as the atlas in the original publication but used a larger dataset and updated minimization algorithms.
The neuropythy.vision.predict_retinotopy function takes one argument, a neuropythy subject object, and yields a pair of dictionaries (lh_data, rh_data) that contain the predicted retinotopy data. The data in the dictionaries are cortical surface properties: they are numpy arrays with one element for each vertex in the subject's relevant hemisphere.
import neuropythy as ny
sub = ny.freesurfer_subject('bert')
(lh_retino, rh_retino) = ny.vision.predict_retinotopy(sub)
tuple([sorted(retino.keys()) for retino in (lh_retino, rh_retino)])
#=> (['angle', 'eccen', 'sigma', 'varea'],
#=> ['angle', 'eccen', 'sigma', 'varea'])
(all(len(v) == sub.lh.vertex_count for v in lh_retino.values()),
all(len(v) == sub.rh.vertex_count for v in rh_retino.values()))
#=> (True, True)The values in the returned dictionaries should be interpreted as follows (but see also neuropythy.vision.as_retinotopy below):
-
'angle'. The polar angle is reported in degrees of rotation around the visual field in the clockwise direction with0°indicating the upper vertical meridian. So+90°is the right horizontal meridian,-90°is the left horizontal meridian, and±180°is the lower vertical meridian. Currently,predict_retinotopyonly returns a symmetric polar angle map, meaning that for both right and left hemispheres, the polar angle will be positive with 0 indicating the UVM and 180 indicating the LVM, but it is possible to set the optional argumentsym_angletoFalsein order to return a full (-180° to 180°) polar angle map. -
'eccen'. The eccentricity is the distance of the pRF center from the fovea, reported in degrees of the visual field. -
'sigma'. The pRF size or radius is the standard deviation of the Gaussian blob used to model the pRF in degrees of visual angle. -
'varea'. The integer visual area label of each vertex; 0 indicates no visual area. The remaining visual areas can be found as follows:import neuropythy as ny mdl = ny.vision.retinotopy_model('benson17', 'lh') # or 'rh' mdl.area_id_to_name #=> pmap({1: 'V1', 2: 'V2', 3: 'V3', 4: 'hV4', 5: 'VO1', #=> 6: 'VO2', 7: 'LO1', 8: 'LO2', 9: 'TO1', 10: 'TO2', #=> 11: 'V3b', 12: 'V3a'})
Once these maps have been generated, they can be saved to disk using ny.save().
Note that, although the keys in the returned dictionary include names like 'angle', these key names are a holdover from much older code and does not conform to neuropythy's typical naming of retinotopy properties; under normal circumstances, these properties would be called 'polar_angle', 'eccentricity', 'radius', and 'visual_area'. This behavior can be fixed, however, by passing the optional argument names='properties'. For more information see the as_retinotopy section below.
An alternative method of predicting retinotopic maps using the Benson et al. (2014) method is to use the benson14_retinotopy command. If you have installed neuropythy, then you can call this command from bash as follows:
# Typical usage:
python -m neuropythy benson14_retinotopy bert
# Detailed options can be seen via -h or --help:
python -m neuropythy benson14_retinotopy --help
The benson14_retinotopy command can be used to project the anatomically defined
template of retinotopy to a subject's left and right hemisphere(s). At least
one subject id (either a freesurfer subject name, if SUBJECTS_DIR is set
appropriately in the environment, or a path to a subject directory) must be
given. In each subject's freesurfer directory, a variety of output data is
deposited:
* surf/lh.angle_benson14 surf/rh.angle_benson14
surf/lh.eccen_benson14 surf/rh.eccen_benson14
surf/lh.varea_benson14 surf/rh.varea_benson14
surf/lh.sigma_benson14 surf/rh.sigma_benson14
These files contain predictions of polar angle, eccentricity, visual-area
label, and pRF radius for each surface vertex in each hemisphere of the
subject's hemispheres. The files are, by default, in FreeSurfer's curv
format, but their format can be modified with the --surf-format flag.
* mri/angle_benson14.mgz
mri/eccen_benson14.mgz
mri/varea_benson14.mgz
mri/sigma_benson14.mgz
These contain the data from the above surface data projected into the
subject's 3D volume. Note that the volumes are oriented like Freesurfer's
mri/brain.mgz file; if you want to convert this to the orientation of your
original anatomical scan, use mri_convert:
> mri_convert -rl mri/rawavg.mgz mri/angle_benson14.mgz \
mri/scanner.angle_benson14.mgz
The following options are accepted:
* --eccen-tag=|-y<tag>
--angle-tag=|-t<tag>
--label-tag=|-l<tag>
--sigma-tag=|-s<tag>
These options specify the output tag to use for the predicted measurement
that results from the registration. By default, these are
'eccen_benson14', 'angle_benson14', 'varea_benson14', and 'sigma_benson14'.
The output files have the name <hemi>.<tag>.mgz
* --surf-format=|-o<nifti|nii.gz|nii|mgh|mgz|curv>
--vol-format=|-v<nifti|nii.gz|nii|mgh|mgz>
These flags specify what format the output should be in; note that nii.gz
and nifti are identical; curv is a FreeSurfer curv (AKA morph data) file.
* --no-volume-export|-x
--no-surface-export|-z
These flags indicate that the various data produced and written to the
filesystem under normal execution should be suppressed. The volume export
refers to the predicted volume files exported to the subject's mri directory
and the surface export refers to the <hemi>.eccen_benson14.mgz and similar
files that are written to the subject's surf directory.
* --subjects-dir=|-d
Specifies additional subject directory search locations (in addition to the
SUBJECTS_DIR environment variable and the FREESURFER_HOME/subjects
directories, which are given here in descending search priority) when looking
for subjects by name. This option cannot be specified multiple times, but it
may contain : characters to separate directories, as in PATH.
* --no-overwrite|-n
This flag indicates that, when writing output files, no file should ever be
replaced, should it already exist.
* --template=|-t
Specifies the specific template that should be applied. By default this is
'Benson17', the 2017 version of the template originally described in the paper
by Benson et al. (2014). The option 'benson14' is also accepted. If the
* --reg=|-R<fsaverage|fsaverage_sym>
Specifies the registration to look for the template in. This is, by default,
fsaverage, but for the templates aligned to the fsaverage_sym hemisphere,
this should specify fsaverage_sym.
* --
This token, by itself, indicates that the arguments that remain should not be
processed as flags or options, even if they begin with a -.Note that a list of subjects may be given after the benson14_retinotopy command name. Each of these may be the path to a FreeSurfer subject's directory or a subject's name, assuming that neuropythy is able to find your FreeSurfer subjects path(s) (see neuropythy configuration for details on telling neuropythy where your subjects are).
The one drawback of the benson14_retinotopy command is that it can only put files in your subject's surf/ and mri/ directories--its output cannot be customized very much. For customized outputs, see the predict_retinotopy method above.
Neuropythy is also capable of making a high-quality estimate of a subject's retinotopic maps based on Bayesian inference. In this operation, the anatomically-defined atlas of retinotopy (see above) is used as a prior of the retinotopic organization while the voxel-wise experimental measurements of retinotopy serve as the observation. This method is described in detail by Benson and Winawer (2018).
To perform Bayesian inference, you must first have measured retinotopic maps for a subject that you have also processed with FreeSurfer:
-
Solve the pRF models for your subjects retinotopy scan(s). As of when this paper was published, we suggest using either VistaSoft (retinotopy tutorial here) or analyzePRF, as these tools yield all the necessary parameters for our tools. See this page for an informal tutorial on how retinotopy might be processed. The specific parameters needed are:
- Polar Angle: rotation around the visual field
- Eccentricity: visual field distance from the fovea
- Sigma/pRF Size: the σ (standard deviation) parameter of the Gaussian pRF
- Variance Explained: the fraction of the BOLD-signal variance explained by the pRF model
-
Project the pRF parameters onto the subject's cortical surface vertices. Note that if you solved for these quantities on the surface (vertices) rather than in the volume (voxels), this step is not necessary. How to project data to the cortical surface is beyond the scope of this wiki, but this tutorial explains how such a transformation can be done with example code (specifically this section), and this tutorial (this section) explains the process we used to create the surface files in this project.
-
Make sure your retinotopy parameter data are in the correct format. In terms of file-formats, it is okay to store your surface data in a FreeSurfer 'curv' format file (also called a 'morph data' file in Python's nibabel library) or either an MGH/MGZ or a NifTI volume file with one non-unitary dimension. More importantly, your retinotopy parameters must be represented in the following ways:
- Polar angle must be in degrees of rotation clockwise starting from the upper vertical meridian. This means that the upper vertical meridian is 0°, the right horizontal meridian is 90°, the left horizontal meridian is -90°, and the lower vertical meridian is ±180°.
- Eccentricity must be in degrees of visual angle from the fovea.
- Sigma/pRF size must be degrees of the visual field.
- Variance explained must be a fraction between v such that 0 ≤ v ≤ 1.
In the following bash code-block, we assume a couple things (make sure to change them if you copy-and-paste!):
- Your subject's FreeSurfer ID is
sub001and your FreeSurferSUBJECTS_DIRis/fs_subjects(so your subject's directory is/fs_subjects/sub001). - You have written out the following surface-data files according to the guidelines above. Note that we assume here you are using MGZ files, but it shouldn't be an issue if you use a different compatible format.
/fs_subjects/sub001/prfs/lh.meas_angle.mgz/fs_subjects/sub001/prfs/lh.meas_eccen.mgz/fs_subjects/sub001/prfs/lh.meas_sigma.mgz/fs_subjects/sub001/prfs/lh.meas_vexpl.mgz/fs_subjects/sub001/prfs/rh.meas_angle.mgz/fs_subjects/sub001/prfs/rh.meas_eccen.mgz/fs_subjects/sub001/prfs/rh.meas_sigma.mgz/fs_subjects/sub001/prfs/rh.meas_vexpl.mgz
- You expect the inferred retinotopy parameters to be written out to the directory
/fs_subjects/sub001/prfs.
To perform the inference, run in a terminal:
python -m neuropythy \
register_retinotopy sub001 \
--verbose \
--surf-outdir=/fs_subjects/sub001/prfs \
--surf-format="mgz" \ # may also be 'curv'
--vol-outdir=/fs_subjects/sub001/prfs \
--vol-format="mgz" \ # may also be 'nifti'
--lh-angle=/fs_subjects/sub001/prfs/lh.meas_angle.mgz \
--lh-eccen=/fs_subjects/sub001/prfs/lh.meas_eccen.mgz \
--lh-radius=/fs_subjects/sub001/prfs/lh.meas_sigma.mgz \
--lh-weight=/fs_subjects/sub001/prfs/lh.meas_vexpl.mgz \
--rh-angle=/fs_subjects/sub001/prfs/rh.meas_angle.mgz \
--rh-eccen=/fs_subjects/sub001/prfs/rh.meas_eccen.mgz \
--rh-radius=/fs_subjects/sub001/prfs/rh.meas_sigma.mgz \
--rh-weight=/fs_subjects/sub001/prfs/rh.meas_vexpl.mgzThis command will run for some time (usually less than 30 minutes) and will produce a set of inferred files (with the same basic data conventions as the input files). In addition, a pair of files (lh.retinotopy.sphere.reg and rh.retinotopy.sphere.reg) which are the subject's cortical surfaces registered to the model of retinotopy. The only other output files that require additional explanation are the varea files (e.g., lh.inferred_varea.mgz). These files contain the visual area labels for each vertex. Any vertex whose label is 0 is not predicted to be part of any visual area, and any inferred parameter for this vertex in the other inferred files should be ignored. For a vertex whose label is not 0, the labels are as in the section on predicting retinotopy above.
Warning: Currently, we have not validated the error or accuracy of areas beyond V1-V3. We include them as a demonstration and guide for future researchers, but we do not endorse the predictions as accurate or valid. Use these predictions at your own risk.
For further information about how to use the Bayesian inference features of neuropythy, visit this OSF wiki page.
Different labs and programs use different conventions when storing and computing over retinotopic maps. Some use cartesian coordinates (x, y) to specify visual field positions while others use polar angle, meaning rotation clockwise in degrees from the upper vertical meridian, and eccentricity; still others might use polar angle but mean rotation counterclockwise in radians from the right horizontal meridian. A few utilities written into neuropythy are designed to help deal with some of these common formats.
One simple tool for extracting retinotopic mapping parameter data from an object with properties or from a dictionary of properties is the neuropythy.retinotopy_data function. Its typical use is to find retinotopy data that is part of a Cortex (subject hemisphere) object and that has a particular prefix.
Example:
>>> import neuropythy as ny
>>> sub = ny.hcp_subject(111312)
# There are many sets of retinotopic properties on some HCP subjects.
>>> [k for k in sub.lh.properties.keys() if k.endswith('eccentricity')]
['prf_eccentricity',
'lowres-prf_eccentricity',
'split1-lowres-prf_eccentricity',
'split2-lowres-prf_eccentricity',
'highres-prf_eccentricity',
'split1-highres-prf_eccentricity',
'split2-highres-prf_eccentricity']
# We can extract a particular one: the dataset starting with 'prf_'.
>>> ny.retinotopy_data(sub.lh, 'prf_')
{'polar_angle': array([...]),
'eccentricity': array([...]),
'radius': array([...]),
'variance_explained': array([...])}Notes:
- The returned retinotopy dictionary is always in neuropythy's
'visual'style (seeas_retinotopy, below); this means that it will always have a'polar_angle'and'eccentricity'property. This means that even if the properties are specified asxandy, the polar angle and eccentricity will be returned. - If a property matching
'variance_explainend'is found, it is included; the same is true of'radius'and'visual_area'. In fact,retinotopy_dataunderstands a variety of common aliases for these properties, which can be examined vianeuropythy.vision.retinotopic_property_aliases. - The second argument (
'prf_'in the example above) can be omitted or can be one of a few special strings:'any','empirical','model', or'none'. In each of these cases but the last, the function will search for any set of retinotopy data with a common set of prefixes; in the middle two cases, only a specific subset of prefixes and suffixes are searched for each case (use of these are deprecated but remain included). If'none'is passed, then only properties such as'polar_angle'(i.e., without any prefix) will be searched.
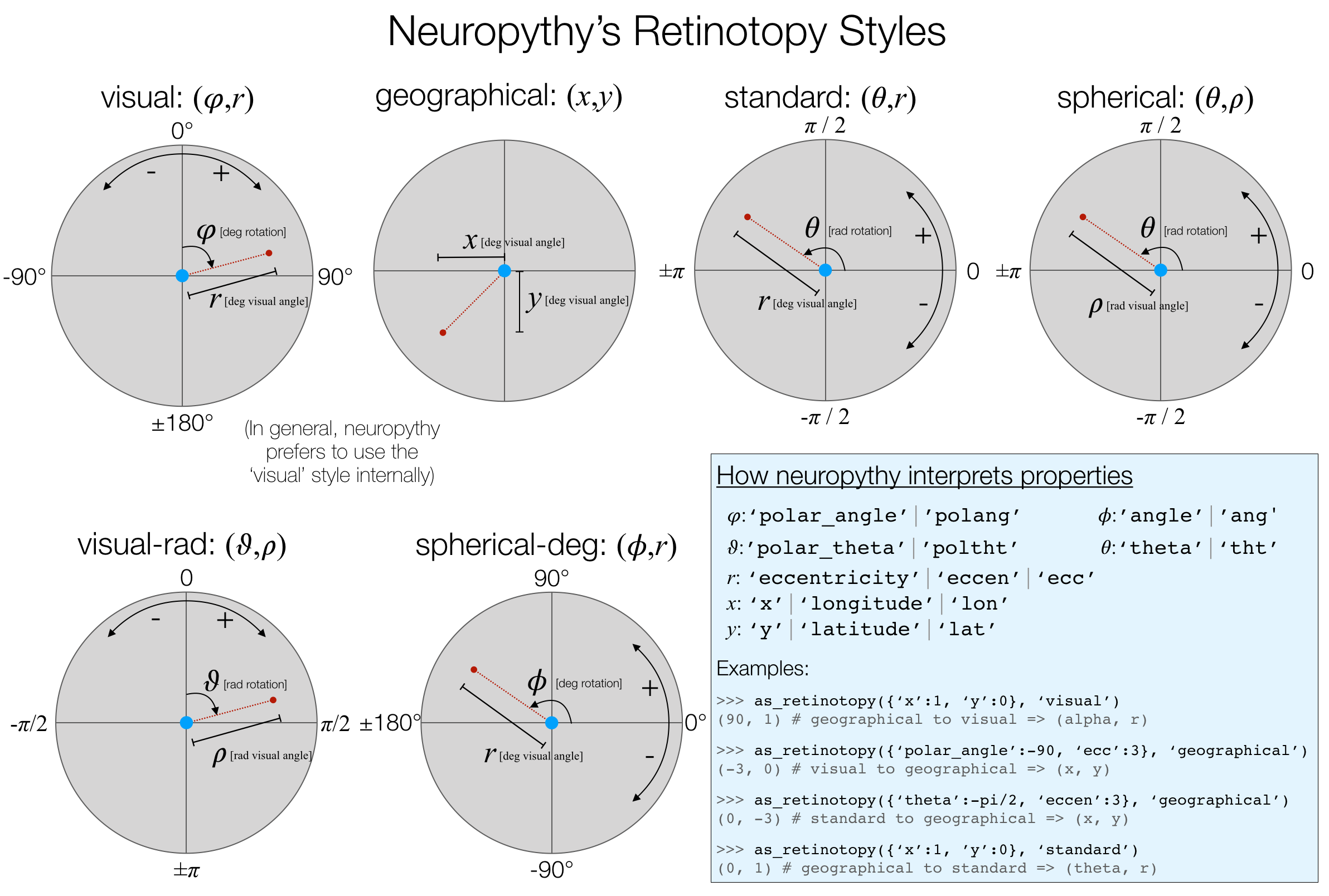
To convert between the various ways of encoding retinotopy, neuropythy understands a set of retinotopy "styles" and can convert between them using the as_retinotopy function. The styles understood by neuropythy are given below:
To use as_retinotopy(), simply pass it a retinotopy dataset (such as that returned by retinotopy_data()) and the name of an output style. The function both examines the property names in the input retinotopy dataset to deduce the input style and converts those data into the output style. The input style is entirely determined by the names of the properties, as shown in the image above.
A critical thing to keep in mind about as_retinotopy is that it always returns a tuple of property values, not a dictionary. So if you request 'geographical' coordinates, it will yield (x,y), and if you request 'visual' you will get (polar_angle, eccentricity).
Smoothing retinotopic maps in the volume or on the cortex is a common technique, especially for visual display of the maps. However, there are reasons to avoid smoothing as well:
- With most kernel-based smoothing, such as the common Gaussian-smoothing, data will always be attenuated away from the extrema of the distribution; for a particular visual area, e.g., V1, this means that V1's data will take up less of the visual field the more they are smoothed, while cortical magnification around the horizontal meridian will grow.
- Many sources of noise, such as vessel artifacts and partial voluming, produce consistent noise in the retinotopic maps that are spatially correlated (i.e., a large blob or smear on the cortical surface is noise). These sources of noise can also yield high-quality fits to pRF models and can be consistent across scans. Such noise is difficult to deal with using traditional smoothing methods and can even be exacerbated by them.
Neuropythy has a built-in function for smoothing retinotopic maps that attempts to get around these problems by minimizing a complicated function of the retinotopic parameters. This function calculates the likelihood of a model that exploits information about the layout of the maps on the cortical surface. Although a complete description of this method is beyond the scope of this wiki-page, a brief overview is given below.
>>> import neuropythy as ny
>>> sub = ny.hcp_subject(111312)
>>> (clean_polang, clean_eccen) = ny.vision.clean_retinotopy(sub.lh, 'prf_')Note that the clean_retinotopy() function is not fast and may require time on the order of hours if run on a high-resolution mesh or using a slower computer.
It is highly recommended that one provide a set of visual area labels (one integer label per vertex) when cleaning retinotopy. The visual area labels may conform to neuropythy's ny.vision.visual_area_names, ny.vision.visual_area_numbers, and ny.vision.visual_area_fieldsigns, or the fieldsigns for each label may be given via an optional argument. As long as a fieldsign can be assigned to each area accurately, the cleaning has considerably more power to accurately smooth the maps.
During cleaning of the retinotopic maps, the routine attempts to minimize the sum of the following terms by moving the mesh vertices around in the visual field:
- A term that gets larger the more each vertex moves away from it's measured/initial pRF position;
- A term that gets larger as the neighboring vertices in the mesh get farther apart in visual space; i.e., this enforces smoothness in space on the cortex;
- A term that is large for triangles in the mesh whose fieldsign does not match that of their vertices' labels and is small when the fieldsign does match;
- A term that gets larger as the difference between the cortical magnification of neighboring triangles in the mesh gets larger.
This method is still somewhat experimental, so its precise default parameters will likely change in the future.
The function documentation for the clean_retinotopy function as well as that of the clean_retinotopy_potential function provide sunstantial additional information if you are trying to learn about the cleaning method. You can see this via help(ny.vision.clean_retinotopy).