-
Notifications
You must be signed in to change notification settings - Fork 2
1.5 BigQuery for Pathogen Detection
Step 1: Go to https://console.cloud.google.com and log into your account using the google account email address you provided to us.
You may have to click through some EULA text.
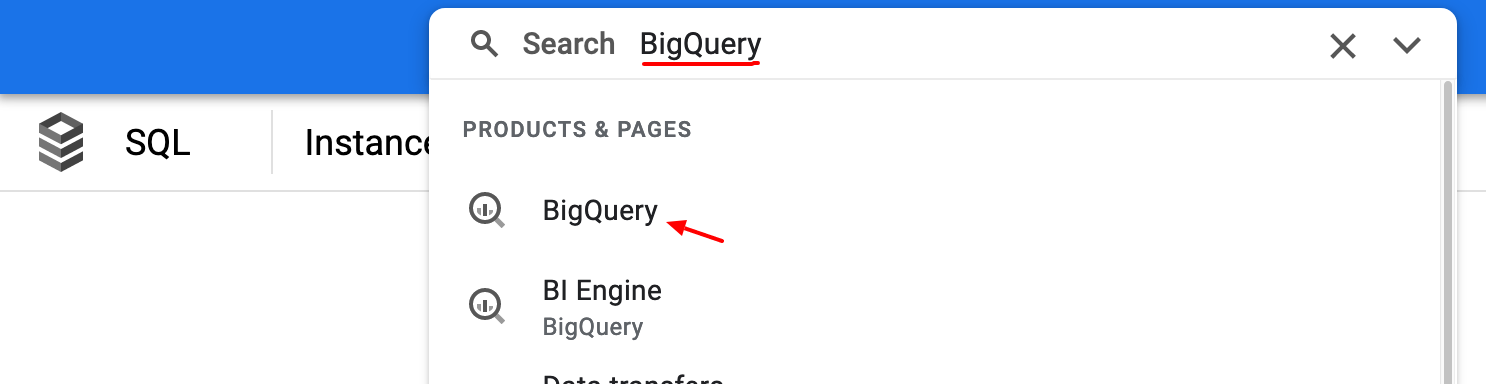 Click the search bar in the top middle of the screen and enter BigQuery as the search term and click on the
BigQuery search result.
Click the search bar in the top middle of the screen and enter BigQuery as the search term and click on the
BigQuery search result.
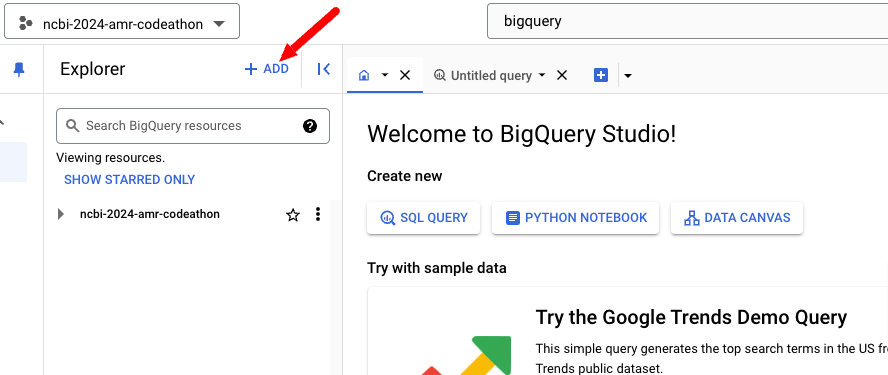
Adding the project ncbi-pathogen-detect to the Data Explorer makes it much
more convenient to get started with the dataset. Click the + ADD button
on the left side of the screen, in the Explorer panel.
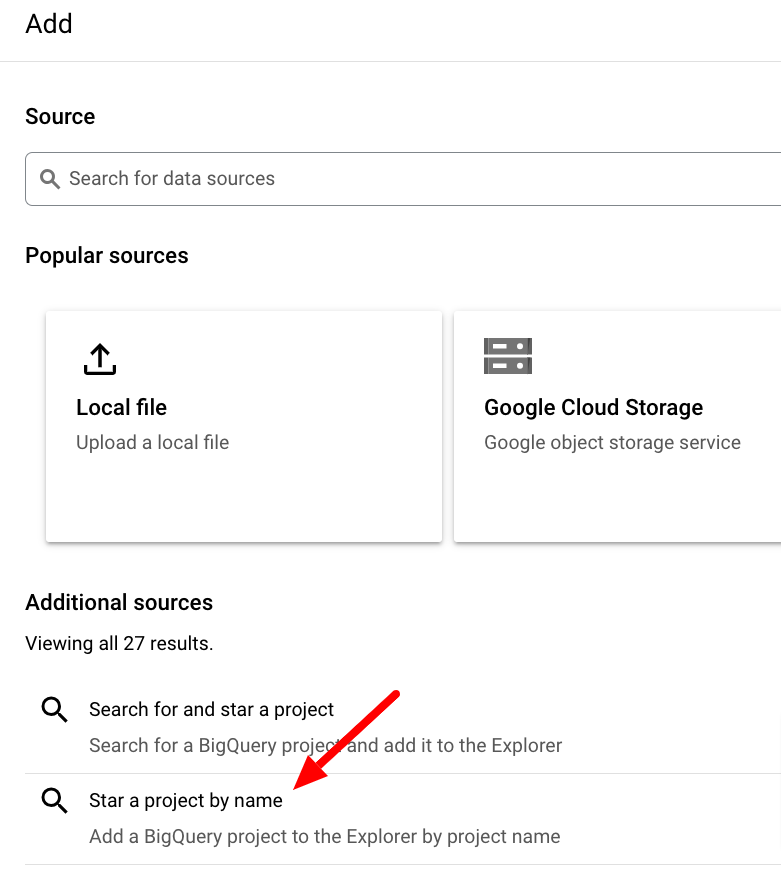
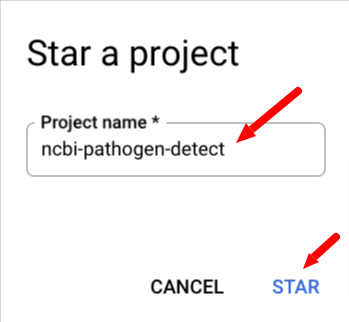
Next, select Star a project by name and paste ncbi-pathogen-detect into
the Pin a project box and click STAR.
You can get to a view of the structure of a table by clicking on the tables in t
he Data Explorer, for example ncbi-pathogen-detect.pdbrowser.microbigge.
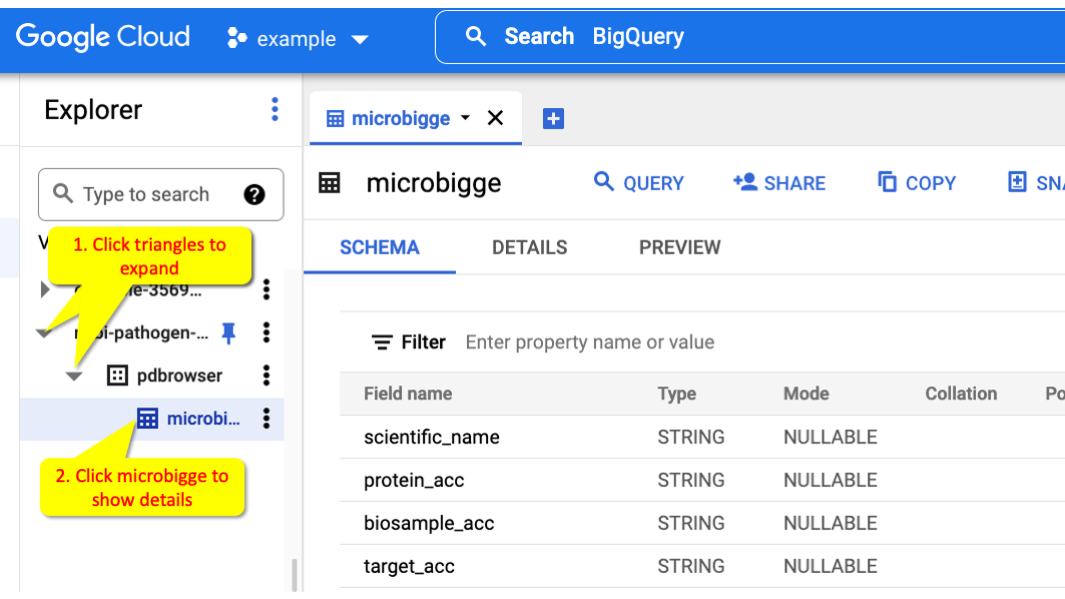
Click the Triangles (1) to expand and show the co
llection. Click the microbigge table (2) to view
information about that table.
Within the table information panel you can click SCHEMA to list the fields a nd PREVIEW to show some sample data from the table.
Click the *Untitled query tab and enter the following query, notice that the table `ncbi-pathogen-detect.pdbrowser.microbigge` is enclosed in backticks (the character under the ~ on the keyboard), then press Run to run the query and get the results.
select count(distinct(biosample_acc)) from `ncbi-pathogen-detect.pdbrowser.microbigge`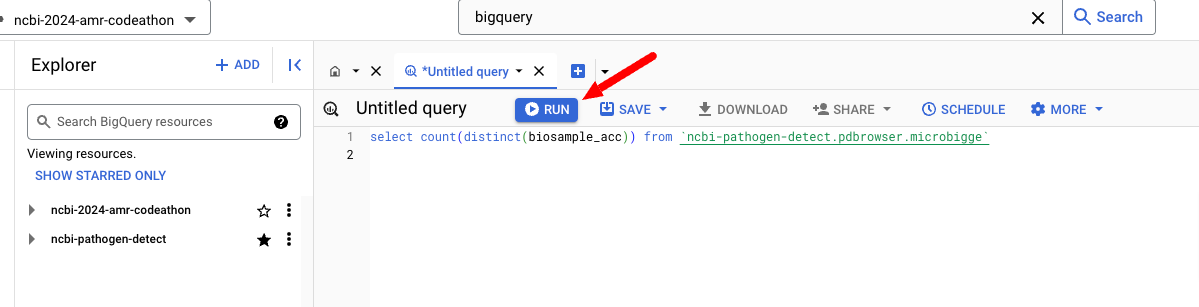
For more information on using BigQuery and SQL see our Useful links page
This work was supported by the National Center for Biotechnology Information of the National Library of Medicine (NLM) and the National Institute of Allergy and Infectious disease (NIAID), National Institutes of Health