-
Notifications
You must be signed in to change notification settings - Fork 1
The Good, the Bad and the Dubious
VHELIBS automatically classifies residues as either 'Good', 'Bad' or 'Dubious' based on a score which depends on some parameters (RSR, RSCC, etc.). But what does that actually mean?
This pages tries to show by example, using the default values, how do they relate to Electron Density Map fitting.
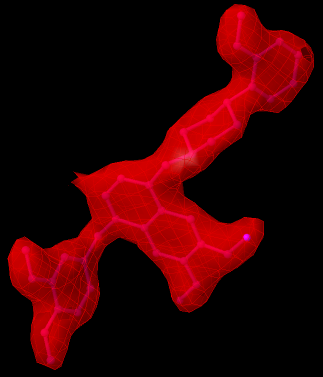
The image above shows a ligand with RSR < 0.24 and RSCC > 0.9, and its electron density map. It can be seen that the ligand model fits reasonably well the EDM. It can be found in the structure 2FVJ.
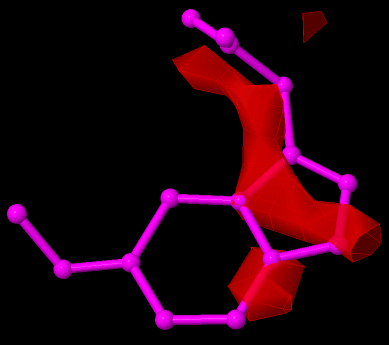
The image above shows a ligands (from 3ADW) with a high RSR (> 0.4) and low (< 0.9) RSCC. As can be seen, it hardly fits the electron density map.
And now the interesting part. Below 3 examples of dubious residues or ligands (with a RSR between 0.24 and 0.4):
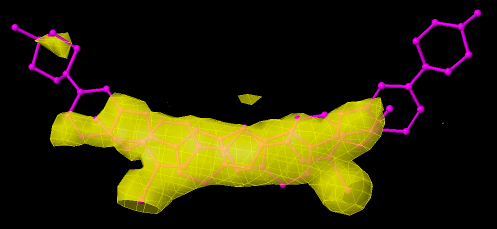
The model of the ligand above (444D) seems to have a problem: there are no EDM for some parts of the molecule and this makes unreliable the conformation of its right end ring. This should be usually classified as Bad. This is a case of a ligand with an average occupancy below 1.0. If we use the default values, (minimum occupancy of 1.0), it will be directly classified as Dubious for that solely. This ligand also has a dubious RSR (between 0.24 and 0.4) and low RSCC (< 0.9), which deems it to be labelled as 'Bad'.
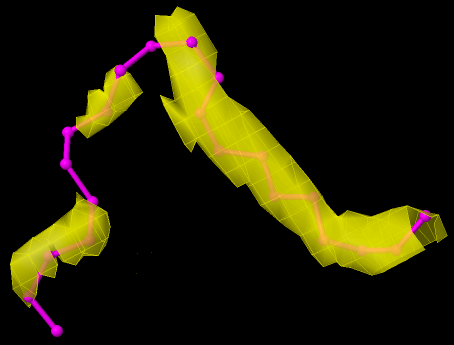
This one (7PRN) is better then the previous one, because although there is no EDM for the whole ligand, the available EDM is enough to confirm the reliability of the ligand coordinates. Therefore, this ligand should be classified as Good, even though it has dubious RSR (between 0.24 and 0.4) and low RSCC (< 0.9).
And finally we have a ligand (3RW0 B4001) which does not perfectly fit the electron density map, but is close enough to it to be marked as Good.
Although in the previous examples mostly ligands have been shown, these principles apply also to the binding sites and any other molecules involved. Here ligands were used for the sake of simplicity.