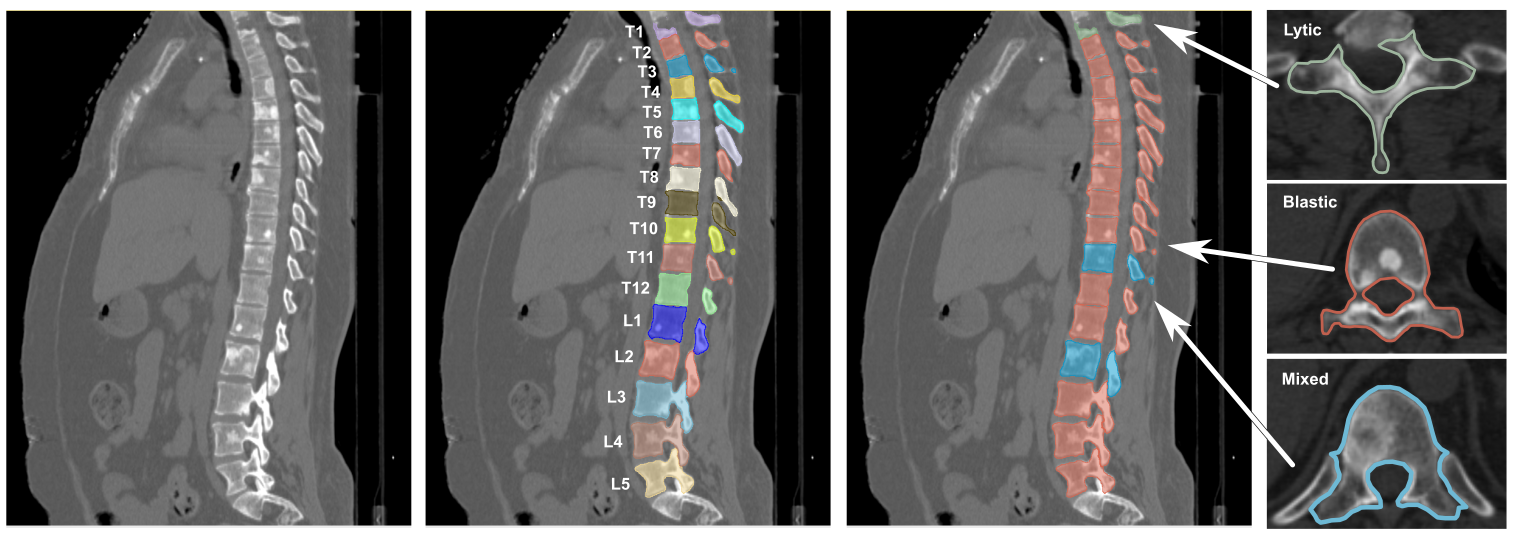
Spine-Mets-CT-SEG: An open annotated dataset and baseline machine learning model for segmentation of vertebrae with metastatic bone lesions from CT.
This repository contains python scripts and model weights to be useed with the Spine-Mets-CT-SEG dataset.
The dataset can be dowloaded here: TCIA Dataset
This collection contains a dataset of 55 CT scans collected on patients with a large range of primary cancers and corresponding bone metastatic lesions obtained for patients with metastatic spine disease. Voxel-level annotations, vertebral level labelling and lesions classification are also provided.
CT scans and segmentations are both provided in DICOM. We recommend 3D Slicer's DICOM viewer for visualization. To correctly import and visualize the segmentations in 3D Slicer the QuantitativeReporting extension is required.
If needed, you can use the script tcia_dcm2nifit.py to convert the full dataset to Nifti file format.
The segmentation weights from the nnUNet training are provided in the folder /weights.
If you use this code or data for your research please cite this paper:
@article{haouchine2024open,
title={An Open Annotated Dataset and Baseline Machine Learning Model for Segmentation of Vertebrae with Metastatic Bone Lesions from CT},
author={Haouchine, Nazim and Hackney, David B and Pieper, Steve D and Wells III, William M and Sanhinova, Malika and Balboni, Tracy A and Spektor, Alexander and Huynh, Mai Ann and Kozono, David E and Doyle, Patrick and others},
journal={medRxiv},
pages={2024--10},
year={2024},
publisher={Cold Spring Harbor Laboratory Press}
}