-
Notifications
You must be signed in to change notification settings - Fork 9
New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
add TSNs from species names using taxize #24
Comments
|
You can also play with a local version of the ITIS database if you want. See the taxize |
|
Cool. Could you give me a line of code that takes species (or higher taxanomy) names and returns TSNs? |
Download the ITIS SQLite database from dropbox to your machinehttps://www.dropbox.com/s/gz1vvsu2d0qps19/itis2_sqlite.zip This DB is 4 months old I think. I had to use a script to convert to a sqlite db, but we could update once the itis site is back up. Install from sql branchinstall_github('taxize_', 'ropensci', 'sql')
library(taxize)Set path to databasetaxize:::taxize_options(localpath = "~/Downloads/itis2.sqlite")You can pass in one or more names to srchkey to get TSNs back.setting returnindex=TRUE gives you back your original search terms so you can parse easily if needed. searchbyscientificname(srchkey=c("oryza sativa","Chironomus riparius","Helianthus annuus","Quercus lobata"), locally=TRUE, returnindex=TRUE)Note that under the cover, these functions are using SQL queries. So you can go in and modify those sql queries if needed. |
|
@schamberlain looks like when treebase does this, it returns several numbers: a tb:identifier.taxon code number, a tb.identifer.taxonVariant code number, a ubio match and a uniprot match: <meta content="11788" datatype="xsd:long" id="meta5204" property="tb:identifier.taxon" xsi:type="nex:LiteralMeta"/>
<meta content="28081" datatype="xsd:long" id="meta5203" property="tb:identifier.taxonVariant" xsi:type="nex:LiteralMeta"/>
<meta href="http://purl.uniprot.org/taxonomy/22658" id="meta5202" rel="skos:closeMatch" xsi:type="nex:ResourceMeta"/>
<meta href="http://www.ubio.org/authority/metadata.php?lsid=urn:lsid:ubio.org:namebank:2651545" id="meta5201" rel="skos:closeMatch" xsi:type="nex:ResourceMeta"/>
<meta href="http://purl.org/phylo/treebase/phylows/study/TB2:S100" id="meta5200" rel="rdfs:isDefinedBy" xsi:type="nex:ResourceMeta"/>(the last meta element refers to the file itself that defines the OTU, which I guess I should add). @rvosa is tb:identifer.taxon the same as the TSN code? The namespace http://purl.org/phylo/treebase/2.0/terms# isn't resolving for me. I suppose it could be useful to have all of these identifiers if we can get them from taxize? I guess the thinking is that at machine that knew one of these identifier vocabularies but not the others could still successfully discover the species covered. Not sure if that is important or not. For the use case of metadata search over a large collection of NeXML files, I am also not sure if it would be worth considering adding other taxonomic hierarchy to the metadata, e.g. facilitating a search for all trees covering order "Passeriformes" without having to look up the order for every OTU in a large collection first. EML files tend to take this approach (giving the full classification). On the other hand, it is redundant and introduces the potential for errors, and a truly fast search should probably index the classification in a database (the way metcat does in EML I think; I guess TreeBase serves much the same function) rather than have to parse every XML file to begin with. Wish I knew more about these issues. Perhaps @rvosa or @hlapp have thoughts on whether it is generally better to make explicit metadata that is already implicit (e.g. given one species id number we can presumably find another algorithmically) or better to minimalist in this? |
|
@schamberlain Sweet! |
|
We may want to think about speed. With very large trees acquiring TSNs could be quite slow if we are calling the ITIS web API. Their API is particularly slow as they only allow one call at a time (e.g. you can't pass in 5 names in one query). NCBI has their own identifiers, and their API is faster. Additionally, my understanding is that ITIS coverage is great if you are in North America, but not so much otherwise. |
|
Personally I would advise against using the TreeBASE vocabulary. The vocabulary is just that - one used by TreeBASE, and not anyone else. Eventually, this would all be covered by the MIAPA ontology, which draws many of its classes and properties from CDAO. The recommendation for TNRS matches is likely also going to be a CDAO property. Furthermore, there is a TNRS vocabulary and example instance documents here: |
|
On Tue, Oct 15, 2013 at 11:40 PM, Carl Boettiger
These things respectively mean the following:
I second Hilmar's suggestion not to use the treebase vocabulary. Sorry that Dr. Rutger A. Vos |
|
@schamberlain Good points about speed, etc. Um, also, looks like the example above is for species names (scientific names) only? Or will it match higher orders? What command would I use that would query both arbitrary taxonomic level and provide fuzzy matching (along with a confidence score, since we can include that in the metadata presumably, though I don't know what the RDFa property would be for that...) Can I get uniprot, ubio, and NCBI identifiers? If NCBI is faster, perhaps we can just use that. @rvosa Thanks for the clarifications. Presumably we can just construct the RDFa given the identifier even if the APIs are not returning RDF responses? @rvosa does it make more sense to provide the identifier in a ReferenceMeta or LiteralMeta (e.g. as full link or just identifier number?) Or maybe just provide both? @ALL I'm guessing there is no definite answer on the explicit vs implicit metadata question. While we can certainly give the user control over what identifiers they want to include and not include as OTU metadata, I think we also want a sensible default that adds, say, one identifier automatically for users who are not familiar with the whole identifier concept to begin with. maybe NCBI is the one to go with? |
|
I agree that it's not disastrous if the response on the other end of a URI Dr. Rutger A. Vos |
|
Okay, I've added the method @rvosa @hlapp Um, I couldn't figure out what Currently we get something that looks like: <otus id="tax1">
<otu label="Struthioniformes" id="t1">
<meta xsi:type="ResourceMeta" href="http://ncbi.nlm.nih.gov/taxonomy/8798" rel="ncbi:id"/>
</otu>
...What other I see that TreeBase trees include a <meta href="http://purl.org/phylo/treebase/phylows/study/TB2:S100" id="meta5200" rel="rdfs:isDefinedBy" xsi:type="nex:ResourceMeta"/>Should we be doing similarly? In general our nexml files won't have a URI. Also, I think I understand the logic of this being related to the ability to reference this otu from another file, but can't we do that without explicitly including such a meta element? I mean, a machine parsing the whole nexml file could obviously generate this line by itself, so I guess it is to allow the otu element to stand alone. But doesn't that become kind of a recursive problem? (e.g. what's the criteria by which a nexml element would need such a line?) |
still needs a real rel property instead of my fake ncbi:id, and needs support for other identifiers
|
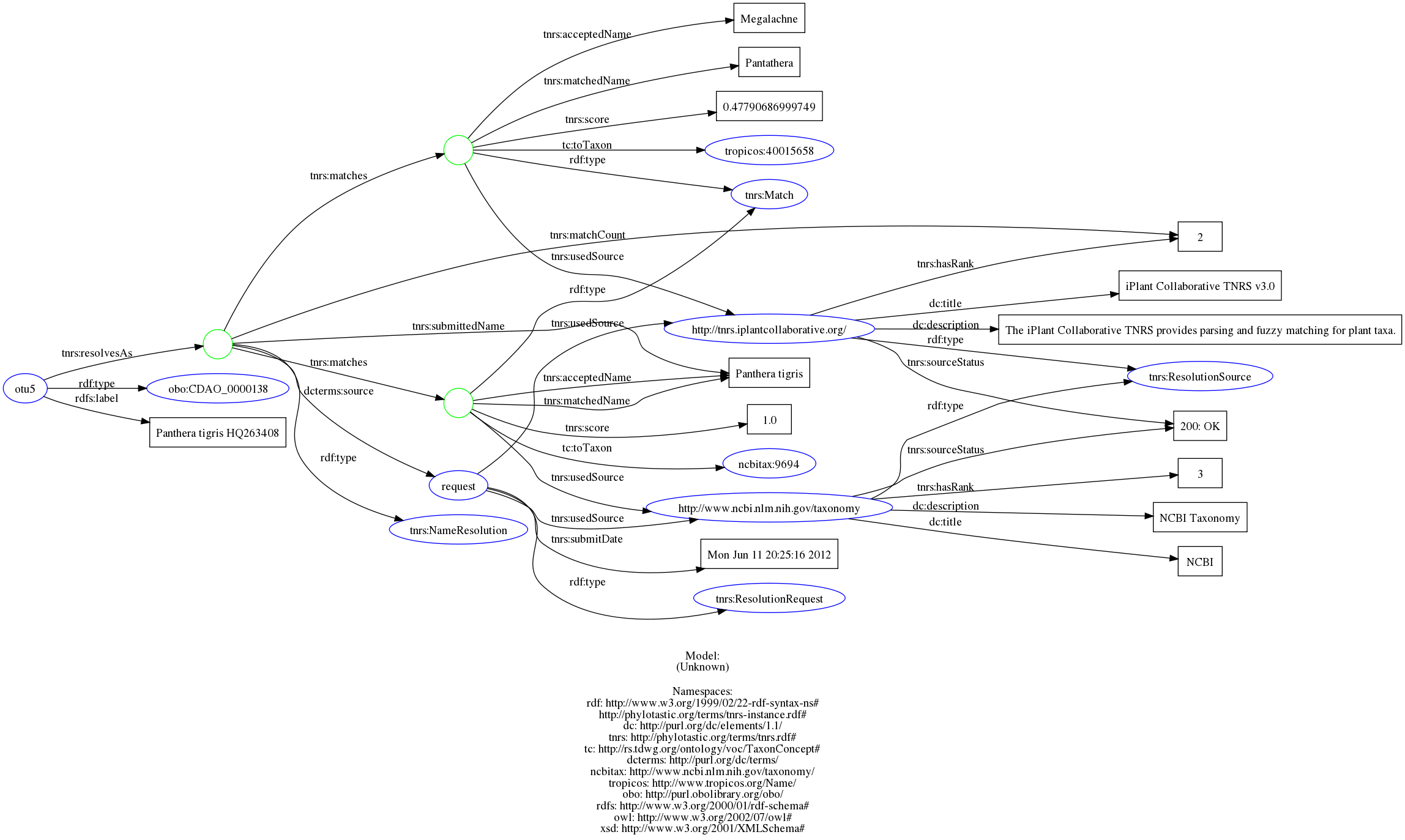
I see... so presumably we could be providing RDFa such as @hlapp illustrates in the TNRS repo annotating each Looks like @hlapp has a handy image showing how much information that crams in: wow. Do we want to include all these triples as annotations to each otu? @sckott I think taxize can provide all the data shown there, though we'd have to generate the meta tagas manually? @ALL for my single NCBI tag, |
|
Re: do you really want all these triples for each OTU: only if you want to preserve full provenance of how the taxon ID assignment was made. So applying taxize to a NeXML object should probably result in that (at least optionally), because presumably taxize at some point has all the information in its hands to fill out those metadata, and the alternative is to throw it all away (not so good for reproducibility in the sense of ability to track provenance of all data). Generally speaking, good functions, and thus good programs don't have side effects. I.e., if I give an input object with all the above metadata to a method that doesn't do anything with taxon ID assignments, the corresponding metadata should reappear in the output unharmed. That said, if the use-case justifies it, you can also stick a taxon ID directly on an OTU. The MIAPA ontology makes the following recommendation for that: <!-- http://rs.tdwg.org/ontology/voc/TaxonConcept#toTaxon -->
<owl:ObjectProperty rdf:about="http://rs.tdwg.org/ontology/voc/TaxonConcept#toTaxon">
<rdfs:comment>For MIAPA reporting, recommended as the property relating an OTU to a taxonomic concept (an entry in a taxonomy, such as an NCBI taxonomy reference) that has been obtained through taxonomic name or other kinds of name resolution or reconciliation procedures.</rdfs:comment>
</owl:ObjectProperty> |
|
Thanks for this, I certainly see your point about documenting the provenance of how we come up with ids if we are going to go and add them programmatically. The use-cases motivating me to add ids in the first place (others may have other use cases) is primarily:
other reasons:
I suppose there is no reason not to include the full provenance, (other than memory perhaps, since R holds the nexml file in working memory at least in the way we do things currently), though it is probably less likely to be of immediate use to the end-user. If I understand your suggestion about how to add a taxon ID directly to an OTU, you suggest the For encoding this into the NeXML, I suppose we can either render this as nested |
Yes, most of it I think. We do have functions interacting with TNRS, NCBI, and Tropicos. Are there other sources we need to pull from? |
|
On Oct 17, 2013, at 5:39 PM, Carl Boettiger wrote:
(Note that with the full provenance you wouldn't have to do even that - you could look at the source TNRS to see where it came from.) That said, if it tuns out as an important use-case to have separate properties for each possible taxon ID source, we can create subproperties. Note, however, that a proliferation of properties really only takes us back to the days of byzantine and idiosyncratic relational schemas where nobody could just agree on how much to normalize things that conceptually are really n-n relationships. The consequence of that is that applications need to understand a bazillion different properties, when instead we could have just inspected the object if we truly needed to know what flavor of a thing it is.
The object value in this case has to be a URI, because it's an entity, not a literal. The object may resolve to RDF, or it may not; in the latter case (or to save a common network roundtrip) we may choose to say more things about it (such as what label it has) directly in the metadata. |
|
@hlapp Thanks. It's a treat to be able to draw on your expertise as I try to wrap my head around semantics better. conceptualYeah, I realize If I understand correctly, ideally the associated I think you're also saying that having a bunch of subproperties is not ideal, since it is unlikely an application understands all of them? practicalOkay, understanding things better but I'm still on the fence as to how we should annotate OTUs to best achieve the 3 use case objectives I mentioned above. (Open to learning about other use cases too). Which of the 3 options below would you recommend we pursue at this stage? Option 1simply gives a link to the NCBI taxon definition using the property (rel) <otus id="tax1">
<otu label="Struthioniformes" id="t1">
<meta xsi:type="ResourceMeta" href="http://ncbi.nlm.nih.gov/taxonomy/8798" rel="tc:toTaxon"/>
</otu>I think this is somewhat analgous to the level of annotation TreeBase provides. Option 2is to add some further annotation to option 1 (not sure what exactly), but not the full provenance. Option 3is to include the full provenance. Not quite sure how that would be expressed. Below, using the example from the TNRS repo I've converted the turtle to RDFa with meta elements, but these aren't valid nex:meta elements since they use While I appreciate the value of having the provenance rather than just displaying the NCBI link in option 1 (with no record to understand where it came from it is arguably worse than just having the taxon <otus id="tax1">
<otu label="Struthioniformes" id="t1">
<meta xmlns:rdf="http://www.w3.org/1999/02/22-rdf-syntax-ns#"
xmlns:rdfs="http://www.w3.org/2000/01/rdf-schema#"
xmlns="http://www.w3.org/1999/xhtml"
xmlns:obo="http://purl.obolibrary.org/obo/"
xmlns:tc="http://rs.tdwg.org/ontology/voc/TaxonConcept#"
xmlns:dc="http://purl.org/dc/elements/1.1/"
xmlns:dcterms="http://purl.org/dc/terms/"
xmlns:tnrs="http://phylotastic.org/terms/tnrs.rdf#"
xmlns:xsd="http://www.w3.org/2001/XMLSchema#"
class="rdf2rdfa">
<meta class="description" about="http://phylotastic.org/terms/tnrs-instance.rdf#otu5"
typeof="obo:CDAO_0000138">
<meta property="rdfs:label" content="Panthera tigris HQ263408"/>
<meta rel="tnrs:resolvesAs">
<meta class="description" typeof="tnrs:NameResolution">
<meta property="tnrs:matchCount" content="2"/>
<meta rel="tnrs:matches">
<meta class="description" typeof="tnrs:Match">
<meta property="tnrs:acceptedName" content="Panthera tigris"/>
<meta property="tnrs:matchedName" content="Panthera tigris"/>
<meta property="tnrs:score" content="1.0"/>
<meta rel="tc:toTaxon" resource="http://www.ncbi.nlm.nih.gov/taxonomy/9694"/>
<meta rel="tnrs:usedSource">
<meta class="description" about="http://www.ncbi.nlm.nih.gov/taxonomy"
typeof="tnrs:ResolutionSource">
<meta property="dc:description" content="NCBI Taxonomy"/>
<meta property="tnrs:hasRank" content="3"/>
<meta property="tnrs:sourceStatus" content="200: OK"/>
<meta property="dc:title" content="NCBI"/>
</meta>
</meta>
</meta>
</meta>
<meta rel="tnrs:matches">
<meta class="description" typeof="tnrs:Match">
<meta property="tnrs:acceptedName" content="Megalachne"/>
<meta property="tnrs:matchedName" content="Pantathera"/>
<meta property="tnrs:score" content="0.47790686999749"/>
<meta rel="tc:toTaxon" resource="http://www.tropicos.org/Name/40015658"/>
<meta rel="tnrs:usedSource">
<meta class="description" about="http://tnrs.iplantcollaborative.org/"
typeof="tnrs:ResolutionSource">
<meta property="dc:description"
content="The iPlant Collaborative TNRS provides parsing and fuzzy matching for plant taxa."/>
<meta property="tnrs:hasRank" content="2"/>
<meta property="tnrs:sourceStatus" content="200: OK"/>
<meta property="dc:title" content="iPlant Collaborative TNRS v3.0"/>
</meta>
</meta>
</meta>
</meta>
<meta rel="dcterms:source">
<meta class="description"
about="http://phylotastic.org/terms/tnrs-instance.rdf#request"
typeof="tnrs:ResolutionRequest">
<meta property="tnrs:submitDate" content="Mon Jun 11 20:25:16 2012"/>
<meta rel="tnrs:usedSource" resource="http://tnrs.iplantcollaborative.org/"/>
<meta rel="tnrs:usedSource" resource="http://www.ncbi.nlm.nih.gov/taxonomy"/>
</meta>
</meta>
<meta property="tnrs:submittedName" content="Panthera tigris"/>
</meta>
</meta>
</meta>
</meta>
</otu> |
|
@cboettig Where are we at on taxonomic ID integration? Any more to be done? I assume so if the issue is still open. Anything to be done on the |
|
It's in there, but just with the minimal annotation to the identifier (like TreeBASE nexml does, not the full provenance). Try: library(RNeXML)
data(bird.orders)
birds <- add_trees(bird.orders)
birds <- taxize_nexml(birds, "NCBI")
nexml_write(birds, "birds.xml")Some draft text now appears at the end of this section: https://github.com/ropensci/RNeXML/blob/devel/inst/doc/pubs/manuscript.md#writing-nexml-metadata |
|
@sckott It would be a good next step if |
|
@cboettig Thanks, will have a look. I don't the best answer off the top for the correct error handling in the RNeXML context. Will have a think about it. |
|
@cboettig Some questions/thoughts:
<meta xsi:type="nex:ResourceMeta" id="m3" source="NCBI" rel="tc:toTaxon"/>
<meta xsi:type="nex:ResourceMeta" id="m3" taxonid="56308" href="http://ncbi.nlm.nih.gov/taxonomy/56308" rel="tc:toTaxon"/>
|
|
@sckott Note that I've started describing this in the manuscript, but not sure how much detail to go into (e.g. detail about NeXML and RDFa that are documented elsewhere, vs detail about RNeXML) so feedback to that end would be great. Yeah, selecting taxon IDs separately and drawing from a mix of authorities for taxonomic annotation makes sense to me; though I'd appreciate perspective from @hlapp on all this. |
|
@sckott: @rvosa raises a good question. What happens if You'll see the function currently just grabs the id returned and throws a warning if it is an na, and otherwise tries to paste it into the metadata: Line 17 in 95ff1e9
Feel free to edit the function to do something more intuitive. @rvosa may have some suggestions. |
|
Okay, since we can now add TSNs from species names using taxize, as this issue stipluates, I think I can close this thread. Further work improving this feature, such as
should be listed under additional issues so they can be organized into appropriate milestones. |
|
Sorry, a bit late to reply. Yes, I'll have a look at the no match found problem. @cboettig Do you prefer those 4 issues you mentioned in one issue (since they are related) or each in separate issues (since they could be solved at different times I guess) What milestone did you have in mind for these taxize issues? or a new one? |
|
@sckott No preference, either one issue or multiple is fine. Yeah, either long-term milestone or some new milestone since I don't think they are critical to the CRAN release (these additions shouldn't break the current API I think) and we probably won't have space to discuss them in the manuscript. (see Current Milestones) |
|
Okay, these moved to the Long Term objectives milestone |
Goal 2 from Kseniia's list, mentioned in #21 (comment)
Will be a good example for the manuscript, once gov't shutdown ends and the taxize servers are back up...
The text was updated successfully, but these errors were encountered: