itree is an interval tree data structure based on a self-balancing AVL binary search tree. Suitable for use with sequence features in bioinformatics.
itree can be installed using pip:
pip install itree
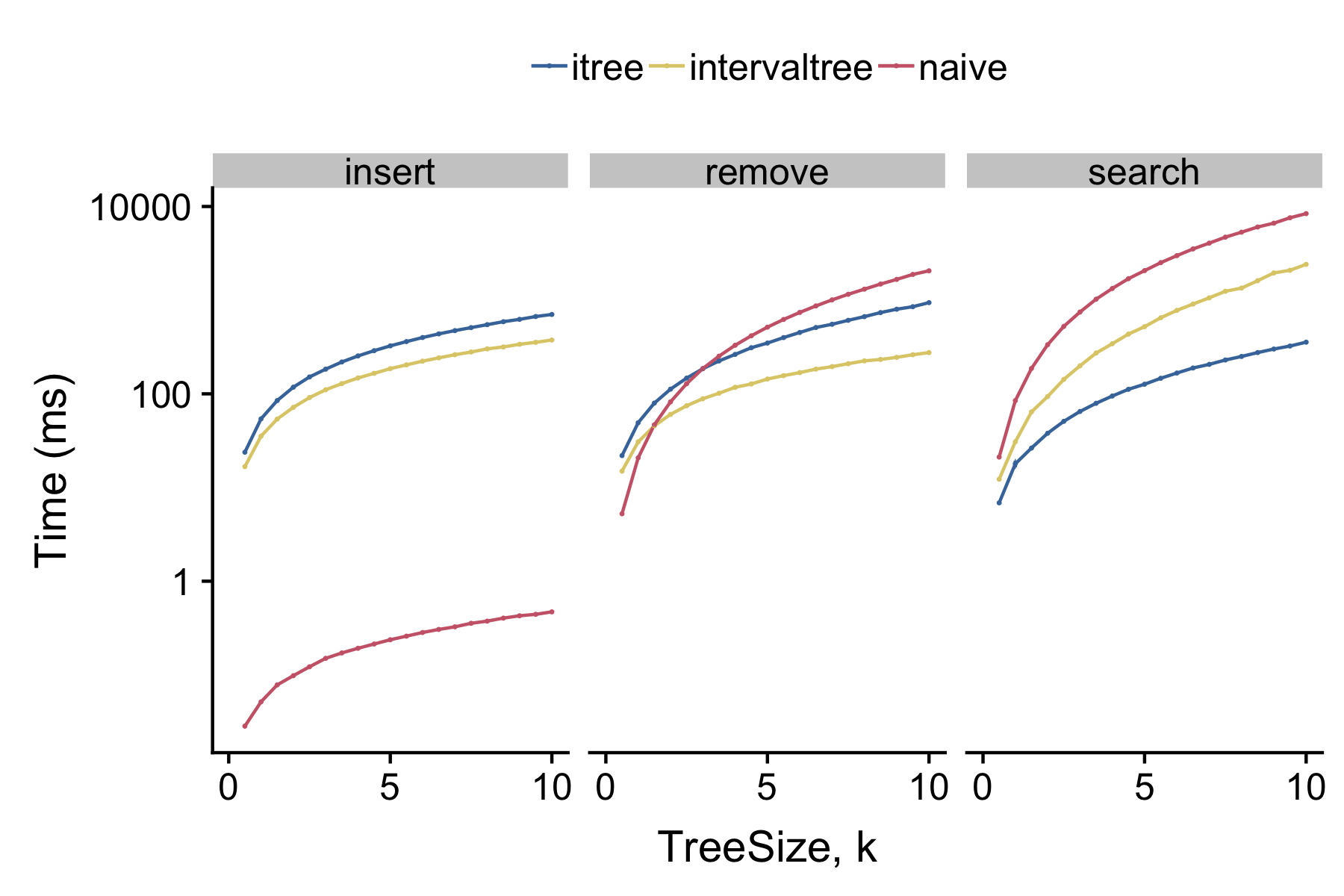
itreeis fast
itree implements an augmented search tree optmized for searching sets of intervals. The following benchmarks the performance of inserting, removing and searching for random intervals taken from the human chromosome 12 Gencode genes[1]:
itreeis convenient
itree has a second-level interface for groups of objects (e.g. chromosomes):
>>> import itree, collections
>>> bed_records = [tuple(l.split()[:3]) for l in open('gencode.bed')]
>>> i = collections.namedtuple('MyInterval', ['chrom','start','end'])
>>> t = itree.GroupedITree('chrom', [i(f[0], int(f[1]), int(f[2])) for f in bed_records])
>>> t.search(i('chr15', 45167200, 45167300))
[MyInterval(chrom='chr15', start=45167213, end=45187956),
MyInterval(chrom='chr15', start=45167250, end=45187952),
MyInterval(chrom='chr15', start=45167213, end=45201175),
MyInterval(chrom='chr15', start=45152663, end=45167526)]- Construction
Creating an interval tree object:
>>> import itree
>>> t = itree.ITree()- Insertion
Any item inserted into an interval tree must contain "start" and "end" attributes as integers.
>>> import collections
>>> i = collections.namedtuple('MyInterval', ['start','end'])
>>> t.insert(i(1,15))
>>> t.insert(i(3,20))
>>> t.insert(i(4,20))
>>> t.insert(i(5,15))
>>> t.insert(i(6,7))- Search
Search for all intervals overlapping a given interval
>>> t.search(i(1,4))
[MyInterval(start=3, end=20), MyInterval(start=4, end=20), MyInterval(start=1, end=15)]- Removal
Remove an interval exactly matching the given interval by its start and end attributes (but not necessarily the
same object).
>>> t.pstring()
┌–(1,15)
–(3,20)
┌–(4,20)
└–(5,15)
└–(6,7)
>>> t.remove(i(1,15))
>>> t.pstring()
┌–(3,20)
└–(4,20)
–(5,15)
└–(6,7)The pstring method is mostly for debugging, but here we illustrate the rebalancing of the tree.
- Grouping
A second-level itree object, GroupedITree, works as a proxy to itree objects which can be grouped by any hashable attribute or function:
>>> import itree, collections
>>> i = collections.namedtuple('Appointment', ['day','start','end'])
>>> appts = [i('Monday', 9, 13), i('Monday', 16, 17), i('Tuesday', 14, 15)]
>>> t = itree.GroupedITree(key='day', intervals=appts)
>>> t.search(i('Monday', 11, 12))
[Appointment(day='Monday', start=9, end=13)]
>>> t.search(i('Monday', 14, 15))
[]You may also use any arbitrary hashable value returned from a function as a key:
>>> i = collections.namedtuple('Appointment', ['day','month','start','end'])
>>> date_key = lambda appt: "{} {}".format(appt.day, appt.month)
>>> appts = [i(5, 'Jan', 9, 13), i(6, 'Jan', 16, 17), i(5, 'Feb', 14, 15)]
>>> t = itree.GroupedITree(key=date_key, intervals=appts)
>>> t.search(i(5, 'Jan', 16, 17))
[]- intervaltree - An interval tree implementation based on a strict binary search tree. Faster insertion and removal but slower search (see above).
- Greeks for geeks explanation of interval trees.
- Advanced data structures - Describes several interval types, including the one min-max interval subset search tree described in section 2.2, which most closely resembles this data structure.
[1] generated with python3 benchmarking/benchmarking.py benchmarking/gencode.chr12.bed 500 10000 500 > benchmarking.txt.