-
Notifications
You must be signed in to change notification settings - Fork 6
Splenocyte
kimmo1019 edited this page Sep 4, 2020
·
7 revisions
git clone https://github.com/kimmo1019/scDEC.git
Downloaded Splenocyte dataset which has been formatted as input of scDEC from the zenode repository. Note that it should be located in the datasets folder.
python main_clustering.py --data Splenocyte --K 12 --dx 8 --dy 20
The results will be saved in the results/Splenocyte folder.
python eval.py --data Splenocyte
The visualization of latent features will be saved as scDEC_embedding.png in the results/Splenocyte folder.
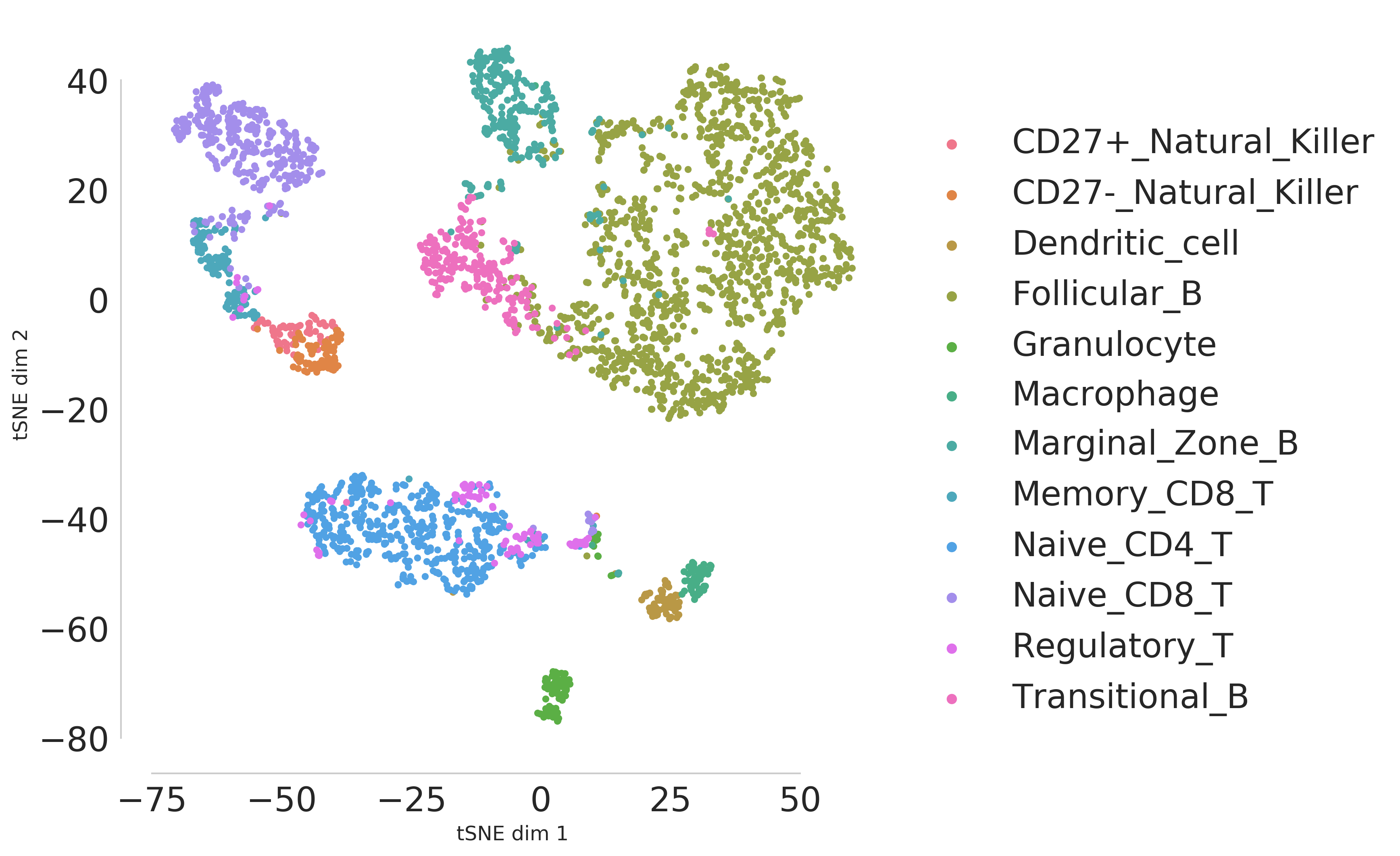
The inferred cluster label will be saved as scDEC_cluster.txt in the same results folder. Each row denotes the predicted cluster id of each cell.
The latent features learned by scDEC will be saved as scDEC_embedding.csv.