-
Notifications
You must be signed in to change notification settings - Fork 19
Forebrain
git clone git://github.com/jsxlei/SCALE.git
cd SCALE
python setup.py install
Get started with downloaded scATAC-seq data Forebrain [Download]
SCALE.py -d Forebrain/data.txt -k 8 --impute
or
SCALE.py -d Forebrain/data.txt -k 8 --binary
input_dir is Forebrain
all results are saved in default output dir output/
Load required packages
import pandas as pd
import numpy as np
from sklearn.metrics import confusion_matrix
from matplotlib import pyplot as plt
import seaborn as sns
from scale.plot import plot_embedding, plot_heatmap
t-SNE embedding is saved in tsne.txt and tsne.pdf labeled by cluster assignments.
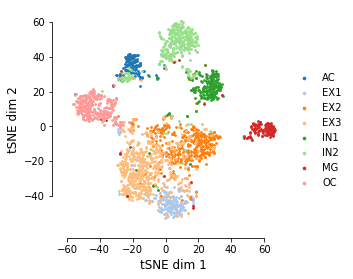
clustering results are saved in cluster_assignments.txt
y = pd.read_csv('output/cluster_assignments.txt', sep='\t', index_col=0, header=None)[1].values
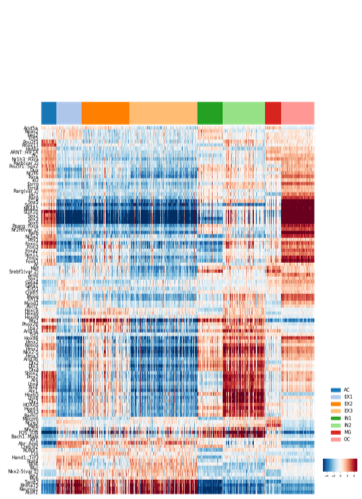
latent feature are saved in feature.txt, we can plot this feature:
feature = pd.read_csv('output/feature.txt', sep='\t', index_col=0, header=None)
plot_heatmap(feature.T, y,
figsize=(8, 3), cmap='RdBu_r', vmax=8, vmin=-8, center=0,
ylabel='Feature dimension', yticklabels=np.arange(10)+1,
cax_title='Feature value', legend_font=6, ncol=1,
bbox_to_anchor=(1.1, 1.1), position=(0.92, 0.15, .08, .04))

imputed data are saved in imputed_data.txt
imputed = pd.read_csv('output/imputed_data.txt', sep='\t', index_col=0)
or binary_imputed folder
from scale.dataset import *
count, peak, barcode = read_mtx('output/binary_imputed')
imputed = pd.DataFrame(count.toarray().T, index=peak, columns=barcode)
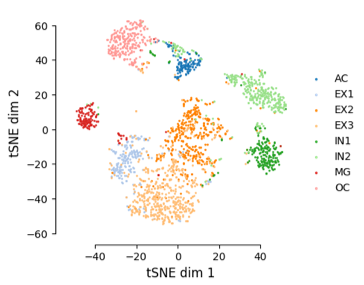
imputed results improved identification of motifs by chromVAR.
left figure is the deviations score of significant motifs(adj_p_value of variability < 0.05).
right figure is the t-SNE plot using the motifs heatmap.
We provide an entropy-based method tto calculate cluster specificity for each peak across cluters.
from scale.specifity import cluster_specific, mat_specificity_score
score_mat = mat_specificity_score(imputed, y)
peak_index, peak_labels = cluster_specific(score_mat, np.unique(y), top=200)
plot_heatmap(imputed.iloc[peak_index], y=y, row_labels=peak_labels, ncol=3, cmap='Reds',
vmax=1, row_cluster=False, legend_font=6, cax_title='Peak Value',
figsize=(8, 10), bbox_to_anchor=(0.4, 1.2), position=(0.8, 0.76, 0.1, 0.015))
