-
Notifications
You must be signed in to change notification settings - Fork 55
Commit
This commit does not belong to any branch on this repository, and may belong to a fork outside of the repository.
[MRG] Replace most RST with Markdown (#973)
* feat: change non-autogen content to use Markdown * update spacing and fix broken link --------- Co-authored-by: dylansdaniels <[email protected]>
- Loading branch information
1 parent
3443b13
commit 5924bb6
Showing
19 changed files
with
1,762 additions
and
1,526 deletions.
There are no files selected for viewing
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| Original file line number | Diff line number | Diff line change |
|---|---|---|
| @@ -0,0 +1,201 @@ | ||
| # hnn-core | ||
|
|
||
| [](https://github.com/jonescompneurolab/hnn-core/actions/?query=branch:master+event:push) | ||
| [](https://circleci.com/gh/jonescompneurolab/hnn-core) | ||
| [](https://codecov.io/gh/jonescompneurolab/hnn-core) | ||
| [](https://pypi.org/project/hnn-core/) | ||
| [](https://gitter.im/jonescompneurolab/hnn-core?utm_source=badge&utm_medium=badge&utm_campaign=pr-badge) | ||
| [](https://doi.org/10.21105/joss.05848) | ||
|
|
||
| 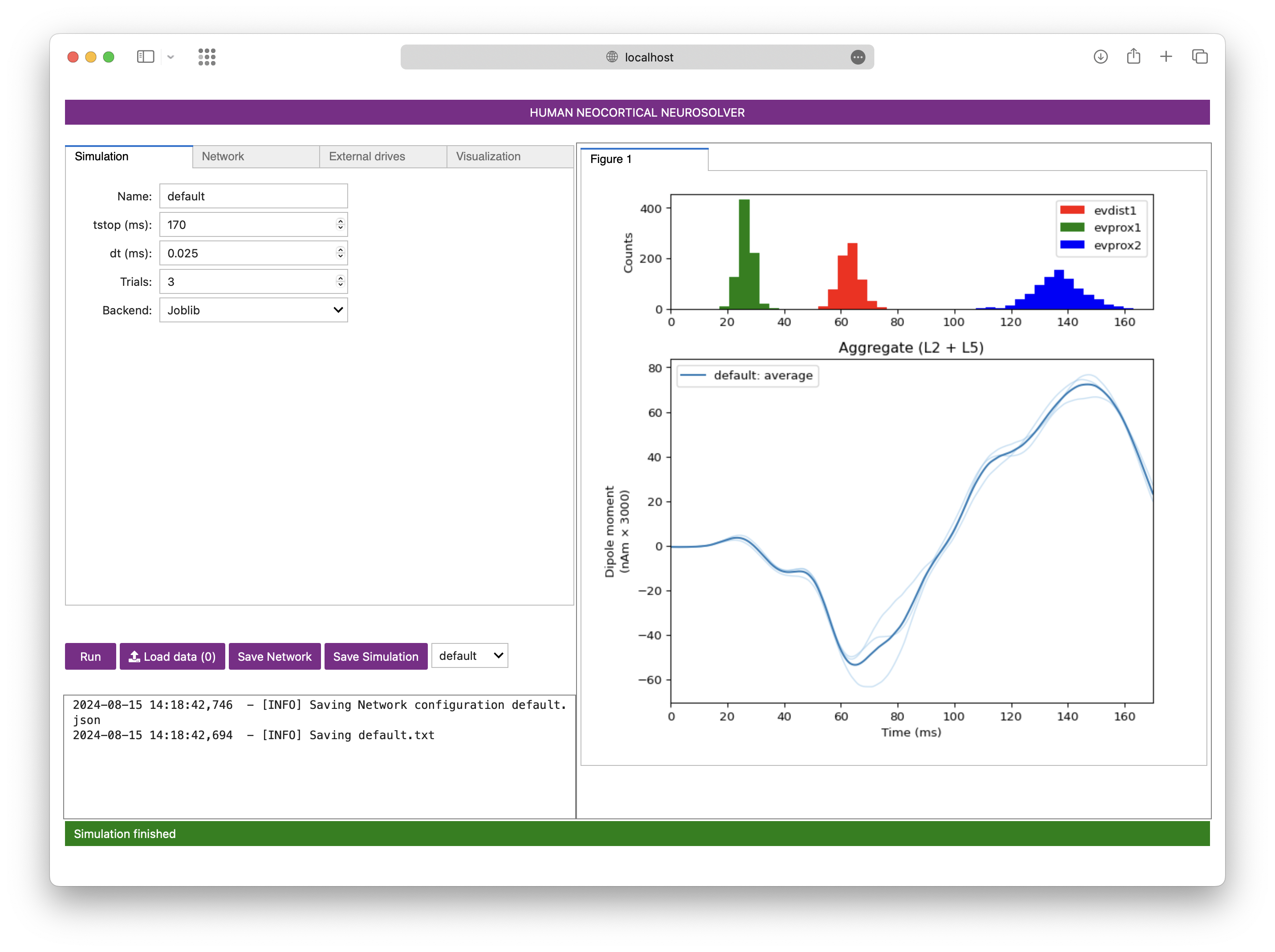 | ||
|
|
||
| ## About | ||
|
|
||
| This is a leaner and cleaner version of the code based off the [HNN | ||
| repository](https://github.com/jonescompneurolab/hnn). | ||
|
|
||
| The **Human Neocortical Neurosolver (HNN)** is an open-source neural modeling | ||
| tool designed to help researchers/clinicians interpret human brain imaging | ||
| data. Based off the original [HNN | ||
| repository](https://github.com/jonescompneurolab/hnn), **HNN-core** provides a | ||
| convenient way to run simulations of an anatomically and biophysically detailed | ||
| dynamical system model of human thalamocortical brain circuits with only a few | ||
| lines of code. Given its modular, object-oriented design, HNN-core makes it | ||
| easy to generate and evaluate hypotheses on the mechanistic origin of signals | ||
| measured with magnetoencephalography (MEG), electroencephalography (EEG), or | ||
| intracranial electrocorticography (ECoG). A unique feature of the HNN model is | ||
| that it accounts for the biophysics generating the primary electric currents | ||
| underlying such data, so simulation results are directly comparable to source | ||
| localized data (current dipoles in units of nano-Ampere-meters); this enables | ||
| precise tuning of model parameters to match characteristics of recorded | ||
| signals. Multimodal neurophysiology data such as local field potential (LFP), | ||
| current-source density (CSD), and spiking dynamics can also be simulated | ||
| simultaneously with current dipoles. | ||
|
|
||
| While the HNN-core API is designed to be flexible and serve users with varying | ||
| levels of coding expertise, the HNN-core GUI is designed to be useful to | ||
| researchers with no formal computational neural modeling or coding experience. | ||
|
|
||
| For more information visit <https://hnn.brown.edu>. There, we describe the use | ||
| of HNN in studying the circuit-level origin of some of the most commonly | ||
| measured MEG/EEG and ECoG signals: event related potentials (ERPs) and low | ||
| frequency rhythms (alpha/beta/gamma). | ||
|
|
||
| Contributors are very welcome. Please read our [contributing | ||
| guide][] if you are interested. | ||
|
|
||
| ## Dependencies | ||
|
|
||
| hnn-core requires Python (>=3.8) and the following packages: | ||
|
|
||
| - numpy | ||
| - scipy | ||
| - matplotlib | ||
| - Neuron (>=7.7) | ||
|
|
||
| ## Optional dependencies | ||
|
|
||
| ### GUI | ||
|
|
||
| - ipywidgets | ||
| - voila | ||
| - ipympl | ||
| - ipykernel | ||
|
|
||
| *Note*: Please follow the **GUI installation** section to install the | ||
| correct GUI dependency versions automatically. | ||
|
|
||
| ### Optimization | ||
|
|
||
| - scikit-learn | ||
|
|
||
| ### Parallel processing | ||
|
|
||
| - joblib (for simulating trials simultaneously) | ||
| - mpi4py (for simulating the cells in parallel for a single trial). | ||
| Also depends on: | ||
| - openmpi or other mpi platform installed on system | ||
| - psutil | ||
|
|
||
| # Installation | ||
|
|
||
| We recommend the [Anaconda Python | ||
| distribution](https://www.anaconda.com/products/individual). To install | ||
| `hnn-core`, simply do: | ||
|
|
||
| $ pip install hnn_core | ||
|
|
||
| and it will install `hnn-core` along with the dependencies which are not | ||
| already installed. | ||
|
|
||
| Note that if you installed Neuron using the traditional installer | ||
| package, it is recommended to remove it first and unset `PYTHONPATH` and | ||
| `PYTHONHOME` if they were set. This is because the pip installer works | ||
| better with virtual environments such as the ones provided by `conda`. | ||
|
|
||
| If you want to track the latest developments of `hnn-core`, you can | ||
| install the current version of the code (nightly) with: | ||
|
|
||
| $ pip install --upgrade https://api.github.com/repos/jonescompneurolab/hnn-core/zipball/master | ||
|
|
||
| To check if everything worked fine, you can do: | ||
|
|
||
| $ python -c 'import hnn_core' | ||
|
|
||
| and it should not give any error messages. | ||
|
|
||
| **Installing optimization dependencies** | ||
|
|
||
| If you are using bayesian optimization, then scikit-learn is required. | ||
| Install hnn-core with scikit-learn using the following command: | ||
|
|
||
| $ pip install hnn_core[opt] | ||
|
|
||
| **GUI installation** | ||
|
|
||
| To install the GUI dependencies along with `hnn-core`, a simple tweak to | ||
| the above command is needed: | ||
|
|
||
| $ pip install hnn_core[gui] | ||
|
|
||
| Note if you are zsh in macOS the command is: | ||
|
|
||
| $ pip install hnn_core'[gui]' | ||
|
|
||
| To start the GUI, please do: | ||
|
|
||
| $ hnn-gui | ||
|
|
||
| **Parallel backends** | ||
|
|
||
| For further instructions on installation and usage of parallel backends | ||
| for using more than one CPU core, refer to our [parallel backend | ||
| guide](https://jonescompneurolab.github.io/hnn-core/stable/parallel.html). | ||
|
|
||
| **Note for Windows users** | ||
|
|
||
| Install Neuron using the [precompiled | ||
| installers](https://nrn.readthedocs.io/en/latest/) **before** installing | ||
| `hnn-core`. Make sure that: | ||
|
|
||
| $ python -c 'import neuron;' | ||
|
|
||
| does not throw any errors before running the install command. If you | ||
| encounter errors, please get help from [NEURON | ||
| forum](https://www.neuron.yale.edu/phpbb/). Finally, do: | ||
|
|
||
| $ pip install hnn_core[gui] | ||
|
|
||
| # Documentation and examples | ||
|
|
||
| Once you have tested that `hnn_core` and its dependencies were | ||
| installed, we recommend downloading and executing the [example | ||
| scripts](https://jonescompneurolab.github.io/hnn-core/stable/auto_examples/index.html) | ||
| provided on the [documentation | ||
| pages](https://jonescompneurolab.github.io/hnn-core/) (as well as in the | ||
| [GitHub repository](https://github.com/jonescompneurolab/hnn-core)). | ||
|
|
||
| Note that `python` plots are by default non-interactive (blocking): each | ||
| plot must thus be closed before the code execution continues. We | ||
| recommend using and 'interactive' python interpreter such as | ||
| `ipython`: | ||
|
|
||
| $ ipython --matplotlib | ||
|
|
||
| and executing the scripts using the `%run`-magic: | ||
|
|
||
| %run plot_simulate_evoked.py | ||
|
|
||
| When executed in this manner, the scripts will execute entirely, after | ||
| which all plots will be shown. For an even more interactive experience, | ||
| in which you execute code and interrogate plots in sequential blocks, we | ||
| recommend editors such as [VS Code](https://code.visualstudio.com) and | ||
| [Spyder](https://docs.spyder-ide.org/current/index.html). | ||
|
|
||
| # Bug reports | ||
|
|
||
| Use the [github issue | ||
| tracker](https://github.com/jonescompneurolab/hnn-core/issues) to report | ||
| bugs. For user questions and scientific discussions, please see our | ||
| [GitHub Discussions | ||
| page](https://github.com/jonescompneurolab/hnn-core/discussions). | ||
|
|
||
| # Interested in Contributing? | ||
|
|
||
| Read our [contributing guide][]. | ||
|
|
||
| # Governance Structure | ||
|
|
||
| Read our [governance | ||
| structure](https://jonescompneurolab.github.io/hnn-core/stable/governance.html). | ||
|
|
||
| # Citing | ||
|
|
||
| If you use HNN-core in your work, please cite our [publication in | ||
| JOSS](https://doi.org/10.21105/joss.05848): | ||
|
|
||
| > Jas et al., (2023). HNN-core: A Python software for cellular and | ||
| > circuit-level interpretation of human MEG/EEG. *Journal of Open Source | ||
| > Software*, 8(92), 5848, <https://doi.org/10.21105/joss.05848> | ||
| [contributing guide]: https://jonescompneurolab.github.io/hnn-core/stable/contributing.html |
Oops, something went wrong.