Hypoxia is a hallmark of solid tumors and a key challenge for cancer therapy. Here we aim to identify the metabolic genes that are critical for tumor’s hypoxic adaptation, and targeting these metabolic genes could selectively inhibiting the proliferation or survival of tumor cells.
The analyses include:
- Comparing the metabolic enzyme network difference between hypoxic and non-hypoxic samples;
- Developed a deep learning model “DepFormer” to predict the hypoxia dependent metabolic genes using lung cancer single cell RNA-seq data, and performed in silico perturbation.
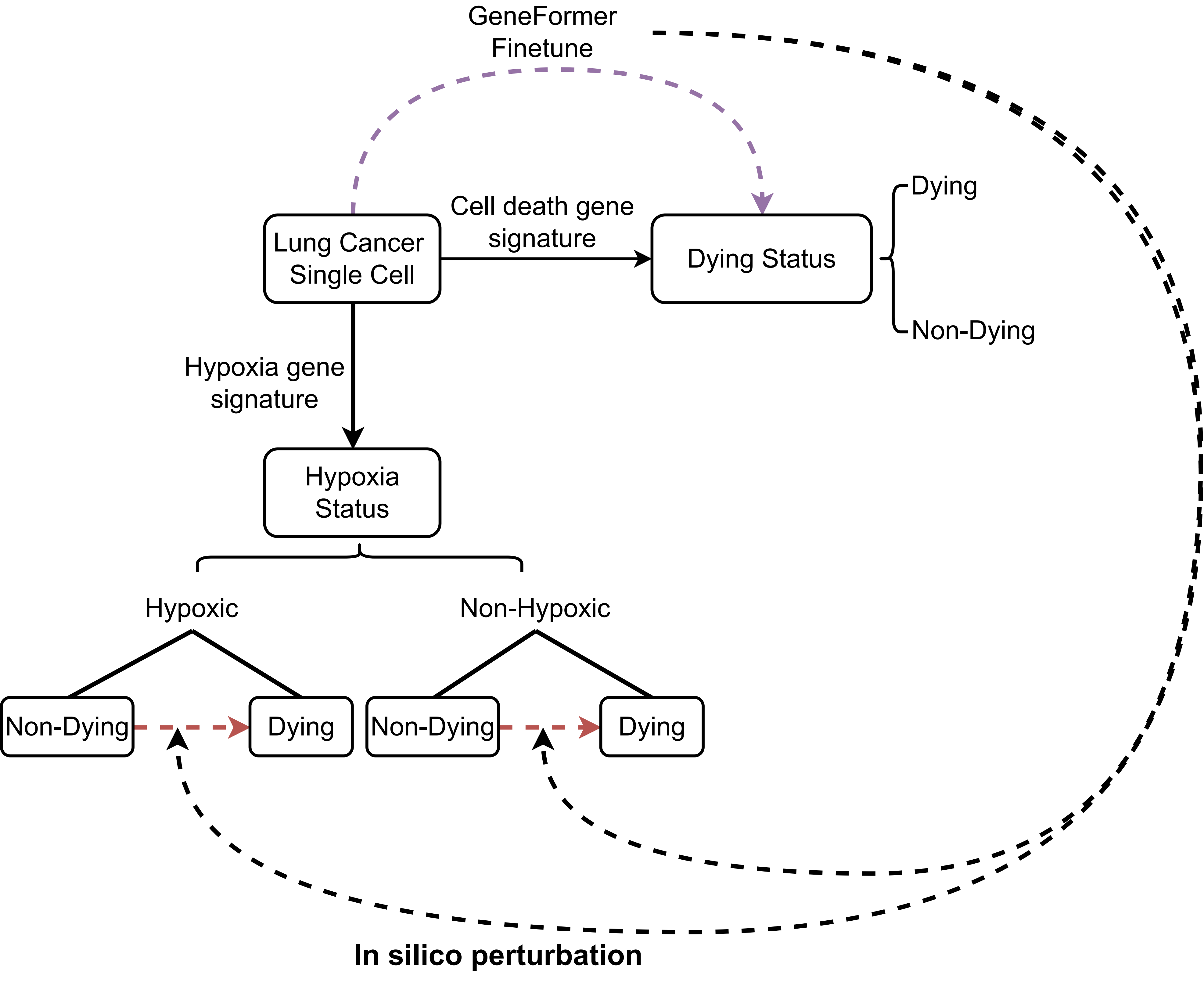
We first defined a cell dying status based on the expression of 12 cell death related gene sets. AUCell was used to calculate the activity score of these gene set for each tumor cell, named cell dying score. The Gaussian mixture model (GMM) is applied to classify these cells into two types: dying cell and non-dying cell. We then fine-tuned the GeneFormer model to learn the dying and non-dying status of cells. To identify genes that are specifically dependent under hypoxic state, we used GMM to divide the cells into hypoxic and non-hypoxic states based on the activity score of hypoxia-related gene sets quantified by AUCell. Subsequently, in silico perturbation was conducted separately in these two cell states. By comparing the results of two situation, we identified potential hypoxia targets. The workflow is:
The code for fine tuning can be found in scripts. The fine-tuned model, corresponding processed training and test data was archived in Zenodo (https://doi.org/10.5281/zenodo.10957878), codes required to reproduce the analysis in this manuscript are available in https://github.com/XSLiuLab/hypoxia_target, and analysis report are available online in https://xsliulab.github.io/hypoxia_target/.
We thank ShanghaiTech University High Performance Computing Public Service Platform for computing services. We thank multi-omics facility, molecular and cell biology core facility of ShanghaiTech University for technical help. This work is supported by cross disciplinary Research Fund of Shanghai Ninth People's Hospital, Shanghai JiaoTong University School of Medicine (JYJC202227). Shanghai Science and Technology Commission (21ZR1442400), National Natural Science Foundation of China (82373149), and startup funding from ShanghaiTech University.
Xiangyu Zhao, Tao Wu & Xue-Song Liu. Deep learning reveals hypoxia vulnerability and FLAD1 as a potential target (Submitted)