-
Notifications
You must be signed in to change notification settings - Fork 81
Home

Announcements: Interested in finding cancer mutations too? Checkout our companion CTAT Mutations Pipeline project.
STAR-Fusion is a component of the Trinity Cancer Transcriptome Analysis Toolkit (CTAT). STAR-Fusion uses the STAR aligner to identify candidate fusion transcripts supported by Illumina reads. STAR-Fusion further processes the output generated by the STAR aligner to map junction reads and spanning reads to a reference annotation set.
Our STAR-Fusion manuscript is published in Genome Biology https://genomebiology.biomedcentral.com/articles/10.1186/s13059-019-1842-9
See STAR-Fusion installation instructions wiki pages.
A short tutorial is available.
There are two ways to run STAR-Fusion. The typical case is that you're staring with FASTQ files. Alternatively, in the context of a more comprehensive transcriptome analysis pipeline leveraging STAR and the human genome from our CTAT genome lib, you may have a 'Chimeric.junction.out' file generated as one of the outputs from an earlier STAR alignment run. If so, you can 'kickstart' STAR-Fusion by using just this 'Chimeric.junction.out' file.
These execution modes are detailed below:
Given paired-end of FASTQ files, run STAR-Fusion like so:
STAR-Fusion --genome_lib_dir /path/to/your/CTAT_resource_lib \
--left_fq reads_1.fq \
--right_fq reads_2.fq \
--output_dir star_fusion_outdir
If you have single-end FASTQ files, just use the --left_fq parameter:
STAR-Fusion --genome_lib_dir /path/to/your/CTAT_resource_lib \
--left_fq reads_1.fq \
--output_dir star_fusion_outdir
If you set the environmental variable 'CTAT_GENOME_LIB' to the '/path/to/your/ctat_genome_lib_build_dir' resulting from the above build process or from the plug-n-play installation, then you won't need to specify --genome_lib_dir as a STAR-Fusion parameter.
Note, unless you have relatively long single-end reads (ex. at least 100 base length), you will be underpowered for detecting fusion transcripts.
If you're running the STAR alignment as part of STAR-Fusion, then you'll need ~40G RAM available. The STAR-Fusion downstream processing doesn't require much RAM.
Alternatively, Kickstart mode: running STAR yourself, and then running STAR-Fusion using the existing outputs
It's not always the case that you want to have STAR-Fusion run STAR directly, as you may have already run STAR earlier on, or prefer to run STAR separately to use the outputs in other processes such as for expression estimates or variant detection.
Parameters that we recommend for running STAR (as of STAR-v2.7.2a) as part of STAR-Fusion are as follows:
STAR --genomeDir ${star_index_dir} \
--readFilesIn ${left_fq_filename} ${right_fq_filename} \
--outReadsUnmapped None \
--twopassMode Basic \
--readFilesCommand "gunzip -c" \
--outSAMstrandField intronMotif \ # include for potential use with StringTie for assembly
--outSAMunmapped Within
Note, the "--twopassMode Basic" setting leads to data-set dependent behavior - ie., if you subsample your reads, there's no guarantee you'll find the same fusion even if the fusion reads are in that subsample. For sample-independent performance, use "--twopassMode None"
and including the following parameters that are relevant to fusion detection and STAR-Fusion execution:
--chimSegmentMin 12 \ # ** essential to invoke chimeric read detection & reporting **
--chimJunctionOverhangMin 8 \
--chimOutJunctionFormat 1 \ # **essential** includes required metadata in Chimeric.junction.out file.
--alignSJDBoverhangMin 10 \
--alignMatesGapMax 100000 \ # avoid readthru fusions within 100k
--alignIntronMax 100000 \
--alignSJstitchMismatchNmax 5 -1 5 5 \ # settings improved certain chimera detections
--outSAMattrRGline ID:GRPundef \
--chimMultimapScoreRange 3 \
--chimScoreJunctionNonGTAG -4 \
--chimMultimapNmax 20 \
--chimNonchimScoreDropMin 10 \
--peOverlapNbasesMin 12 \
--peOverlapMMp 0.1 \
--alignInsertionFlush Right \
--alignSplicedMateMapLminOverLmate 0 \
--alignSplicedMateMapLmin 30
Essential parameters for use with STAR-Fusion are indicted. Others are current settings used when the alignment is performed with STAR-Fusion.
This will (in part) generate a file called 'Chimeric.out.junction', which is used by STAR-Fusion like so:
STAR-Fusion --genome_lib_dir /path/to/your/CTAT_resource_lib \
-J Chimeric.out.junction \
--output_dir star_fusion_outdir
Note, if you use this -J Chimeric.out.junction file, it is essential that the new '--chimOutJunctionFormat 1' parameter was used, since this integrates alignment statistics at the end of the junction file, which are used for estimating the FFPM fusion ~expression estimates.
The output from STAR-Fusion is found as a tab-delimited file named 'star-fusion.fusion_predictions.tsv', along with an abridged version that excludes the identification of the evidence fusion reads and called 'star-fusion.fusion_predictions.abridged.tsv', with the following format:
#FusionName JunctionReadCount SpanningFragCount SpliceType LeftGene LeftBreakpoint RightGene RightBreakpoint LargeAnchorSupport FFPM LeftBreakDinuc LeftBreakEntropy RightBreakDinuc RightBreakEntropy annots
THRA--AC090627.1 27 93 ONLY_REF_SPLICE THRA^ENSG00000126351.8 chr17:38243106:+ AC090627.1^ENSG00000235300.3 chr17:46371709:+ YES_LDAS 23875.8456 GT 1.8892 AG 1.9656 ["CCLE","FA_CancerSupp","INTRACHROMOSOMAL[chr17:8.12Mb]"]
THRA--AC090627.1 5 93 ONLY_REF_SPLICE THRA^ENSG00000126351.8 chr17:38243106:+ AC090627.1^ENSG00000235300.3 chr17:46384693:+ YES_LDAS 19498.6072 GT 1.8892 AG 1.4295 ["CCLE","FA_CancerSupp","INTRACHROMOSOMAL[chr17:8.12Mb]"]
ACACA--STAC2 12 52 ONLY_REF_SPLICE ACACA^ENSG00000132142.15 chr17:35479453:- STAC2^ENSG00000141750.6 chr17:37374426:- YES_LDAS 12733.7844 GT 1.9656 AG 1.9656 ["ChimerSeq","CCLE","Klijn_CellLines","FA_CancerSupp","INTRACHROMOSOMAL[chr17:1.60Mb]"]
RPS6KB1--SNF8 10 43 ONLY_REF_SPLICE RPS6KB1^ENSG00000108443.9 chr17:57970686:+ SNF8^ENSG00000159210.5 chr17:47021337:- YES_LDAS 10545.1651 GT 1.3753 AG 1.8323 ["Klijn_CellLines","FA_CancerSupp","ChimerSeq","CCLE","INTRACHROMOSOMAL[chr17:10.95Mb]"]
TOB1--SYNRG 8 30 ONLY_REF_SPLICE TOB1^ENSG00000141232.4 chr17:48943419:- SYNRG^ENSG00000006114.11 chr17:35880751:- YES_LDAS 7560.6844 GT 1.4566 AG 1.8892 ["FA_CancerSupp","CCLE","INTRACHROMOSOMAL[chr17:12.97Mb]"]
VAPB--IKZF3 4 46 ONLY_REF_SPLICE VAPB^ENSG00000124164.11 chr20:56964573:+ IKZF3^ENSG00000161405.12 chr17:37934020:- YES_LDAS 9948.269 GT 1.9656 AG 1.7819 ["FA_CancerSupp","Klijn_CellLines","CCLE","ChimerSeq","ChimerPub","INTERCHROMOSOMAL[chr20--chr17]"]
ZMYND8--CEP250 2 44 ONLY_REF_SPLICE ZMYND8^ENSG00000101040.15 chr20:45852970:- CEP250^ENSG00000126001.11 chr20:34078463:+ NO_LDAS 9152.4075 GT 1.8295 AG 1.8062 ["FA_CancerSupp","CCLE","ChimerSeq","INTRACHROMOSOMAL[chr20:11.74Mb]"]
AHCTF1--NAAA 3 38 ONLY_REF_SPLICE AHCTF1^ENSG00000153207.10 chr1:247094880:- NAAA^ENSG00000138744.10 chr4:76846964:- YES_LDAS 8157.5805 GT 1.7232 AG 1.8062 ["FA_CancerSupp","CCLE","INTERCHROMOSOMAL[chr1--chr4]"]
VAPB--IKZF3 1 46 ONLY_REF_SPLICE VAPB^ENSG00000124164.11 chr20:56964573:+ IKZF3^ENSG00000161405.12 chr17:37922746:- NO_LDAS 9351.3729 GT 1.9656 AG 1.9329 ["FA_CancerSupp","Klijn_CellLines","CCLE","ChimerSeq","ChimerPub","INTERCHROMOSOMAL[chr20--chr17]"]
VAPB--IKZF3 1 46 ONLY_REF_SPLICE VAPB^ENSG00000124164.11 chr20:56964573:+ IKZF3^ENSG00000161405.12 chr17:37944627:- NO_LDAS 9351.3729 GT 1.9656 AG 1.8892 ["FA_CancerSupp","Klijn_CellLines","CCLE","ChimerSeq","ChimerPub","INTERCHROMOSOMAL[chr20--chr17]"]
STX16--RAE1 4 33 ONLY_REF_SPLICE STX16^ENSG00000124222.17 chr20:57227143:+ RAE1^ENSG00000101146.8 chr20:55929088:+ YES_LDAS 7361.719 GT 1.9899 AG 1.9656 ["FA_CancerSupp","CCLE","INTRACHROMOSOMAL[chr20:1.27Mb]"]
AHCTF1--NAAA 1 38 ONLY_REF_SPLICE AHCTF1^ENSG00000153207.10 chr1:247094431:- NAAA^ENSG00000138744.10 chr4:76846964:- NO_LDAS 7759.6498 GT 1.9086 AG 1.8062 ["FA_CancerSupp","CCLE","INTERCHROMOSOMAL[chr1--chr4]"]
STX16-NPEPL1--RAE1 4 24 INCL_NON_REF_SPLICE STX16-NPEPL1^ENSG00000254995.4 chr20:57227143:+ RAE1^ENSG00000101146.8 chr20:55929088:+ YES_LDAS 5571.0306 GT 1.9899 AG 1.9656 INTRACHROMOSOMAL[chr20:1.27Mb]
RAB22A--MYO9B 6 11 ONLY_REF_SPLICE RAB22A^ENSG00000124209.3 chr20:56886178:+ MYO9B^ENSG00000099331.9 chr19:17256207:+ YES_LDAS 3382.4115 GT 1.6895 AG 1.9656 ["FA_CancerSupp","ChimerSeq","CCLE","INTERCHROMOSOMAL[chr20--chr19]"]
MED1--ACSF2 4 11 ONLY_REF_SPLICE MED1^ENSG00000125686.7 chr17:37595418:- ACSF2^ENSG00000167107.8 chr17:48548389:+ YES_LDAS 2984.4807 GT 1.9656 AG 1.9656 ["FA_CancerSupp","CCLE","INTRACHROMOSOMAL[chr17:10.90Mb]"]
MED13--BCAS3 2 12 ONLY_REF_SPLICE MED13^ENSG00000108510.5 chr17:60129898:- BCAS3^ENSG00000141376.16 chr17:59469338:+ YES_LDAS 2785.5154 GT 1.5546 AG 1.9086 ["FA_CancerSupp","CCLE","INTRACHROMOSOMAL[chr17:0.55Mb]"]
MED1--STXBP4 1 15 ONLY_REF_SPLICE MED1^ENSG00000125686.7 chr17:37607291:- STXBP4^ENSG00000166263.9 chr17:53218671:+ NO_LDAS 3183.4461 GT 1.3996 AG 1.7968 ["CCLE","FA_CancerSupp","Klijn_CellLines","INTRACHROMOSOMAL[chr17:15.44Mb]"]
MED13--BCAS3 1 12 ONLY_REF_SPLICE MED13^ENSG00000108510.5 chr17:60129898:- BCAS3^ENSG00000141376.16 chr17:59465979:+ NO_LDAS 2586.55 GT 1.5546 AG 0.8366 ["FA_CancerSupp","CCLE","INTRACHROMOSOMAL[chr17:0.55Mb]"]
STARD3--DOK5 2 7 ONLY_REF_SPLICE STARD3^ENSG00000131748.11 chr17:37793484:+ DOK5^ENSG00000101134.7 chr20:53259997:+ NO_LDAS 1790.6885 GT 1.8892 AG 1.9656 ["FA_CancerSupp","CCLE","INTERCHROMOSOMAL[chr17--chr20]"]
DIDO1--TTI1 1 10 ONLY_REF_SPLICE DIDO1^ENSG00000101191.12 chr20:61569148:- TTI1^ENSG00000101407.8 chr20:36642259:- NO_LDAS 2188.6192 GT 1.6402 AG 1.9329 ["FA_CancerSupp","ChimerSeq","CCLE","INTRACHROMOSOMAL[chr20:24.85Mb]"]
DIDO1--TTI1 1 10 ONLY_REF_SPLICE DIDO1^ENSG00000101191.12 chr20:61569148:- TTI1^ENSG00000101407.8 chr20:36634799:- NO_LDAS 2188.6192 GT 1.6402 AG 1.8892 ["FA_CancerSupp","ChimerSeq","CCLE","INTRACHROMOSOMAL[chr20:24.85Mb]"]
BRD4--RFX1 1 8 ONLY_REF_SPLICE BRD4^ENSG00000141867.13 chr19:15443101:- RFX1^ENSG00000132005.4 chr19:14109129:- NO_LDAS 1790.6884 GT 1.9086 AG 1.8892 ["CCLE","FA_CancerSupp","INTRACHROMOSOMAL[chr19:1.23Mb]"]
BRD4--RFX1 1 8 ONLY_REF_SPLICE BRD4^ENSG00000141867.13 chr19:15443101:- RFX1^ENSG00000132005.4 chr19:14094407:- NO_LDAS 1790.6884 GT 1.9086 AG 1.8295 ["CCLE","FA_CancerSupp","INTRACHROMOSOMAL[chr19:1.23Mb]"]
TRPC4AP--MRPL45 1 8 ONLY_REF_SPLICE TRPC4AP^ENSG00000100991.7 chr20:33665849:- MRPL45^ENSG00000174100.5 chr17:36478009:+ NO_LDAS 1790.6884 GT 1.6895 AG 1.9086 ["CCLE","Klijn_CellLines","FA_CancerSupp","INTERCHROMOSOMAL[chr20--chr17]"]
The JunctionReads column indicates the number of RNA-Seq fragments containing a read that aligns as a split read at the site of the putative fusion junction.
The SpanningFrags column indicates the number of RNA-Seq fragments that encompass the fusion junction such that one read of the pair aligns to a different gene than the other paired-end read of that fragment.
Those predictions that have very few JunctionReads and/or SpanningReads are going to be enriched for false positives. Note, depending on the site of the fusion breakpoint and length of the reads, it may not be possible to have SpanningFragments and all evidence may show up in the form of JunctionReads.
The number of fusion-supporting reads depends on both the expression of the fusion transcript and the number of reads sequenced. The deeper the sequenced data set, the greater the number of artifactual fusions that will appear with minimal supporting evidence, and so taking into account the sequencing depth is important to curtail overzealous prediction of fusion transcripts with ever-so-minimal supporting evidence. We provide normalized measures of the fusion-supporting rna-seq fragments as FFPM (fusion fragments per million total reads) measures. A filter of 0.1 sum FFPM (meaning at least 1 fusion-supporting rna-seq fragment per 10M total reads) tends to be effective at excluding fusion artifacts, and is the current default for filtering fusions from the final output. Adjust the 'STAR-Fusion --min_FFPM' parameter, or set it to zero to disable FFPM-based filtering.
The 'LargeAnchorSupport' column indicates whether there are split reads that provide 'long' (set to length of 25 bases) alignments on both sides of the putative breakpoint. Those fusions supported only by split reads (no spanning fragments) and lack LargeAnchorSupport are often highly suspicious and tend to be false positives. Those with LargeAnchorSupport are labeled as 'YES_LDAS' (where LDAS = long double anchor support.... yes, more jargon).
'SpliceType' indicates whether the proposed breakpoint occurs at reference exon junctions as provided by the reference transcript structure annotations (ex. gencode).
'LeftBreakEntropy' and 'RightBreakEntropy' represent the Shannon entropy of the 15 exonic bases flanking the breakpoint. The maximum entropy is 2, representing highest complexity. The lowest would be zero (involving a 15 base mononucleotide run). Low entropy sites should generally be treated as less confident breakpoints.
The abridged output file contents are shown above. See the unabridged 'star-fusion.fusion_predictions.tsv' output file for the identity of the RNA-Seq fragments identified as junction or spanning fragments, where the individual read names are provided as comma-delimited lists in each corresponding column.
The final column 'annots' provides a simplified annotation for fusion transcript, leveraging FusionAnnotator (bundled with STAR-Fusion). For the human source or plug-n-play genome libs, the fusion annotation info is based on CTAT_HumanFusionLib, which includes many popular resources for annotating fusions known to be relevant to cancer, as well as fusions thought to be red herrings that will be automatically filtered from the final output. Rules for filtering out fusions based on annotations are encoded in a small Perl module found as '${CTAT_GENOME_LIB}/AnnotFilterRule.pm', and for the provided human genome lib involves excluding 'red herring' categories (described here - not a fish picture ;-) ) in addition to fusions involving mitochondrial genes or HLA loci (common artifacts). To exclude any annotation-based filtering, use the 'STAR-Fusion --no_annotation_filter' parameter.
If there are alternatively spliced isoforms for fusion transcripts, the same fusion pair will be listed as multiple entries but with different breakpoints identified.
We have a companion tool called FusionInspector that provides a more in-depth view of the evidence supporting the predicted fusions. FusionInspector can also run Trinity to de novo reconstruct your predicted fusion transcripts based on the identified fusion-supporting RNA-Seq reads.
As of STAR-Fusion v1.1.0, FusionInspector is integrated into STAR-Fusion as a submodule.
FusionInspector can be run in either 'inspect' or 'validate' mode when executed downstream from STAR-Fusion:
-
'--FusionInspector inspect': only the reads identified by STAR-Fusion as evidence supporting the fusion prediction are aligned directly to a target set of fusion-gene contigs for exploration using IGV.
-
'--FusionInspector validate': involves a more rigorous process of reevaluating the entire set of input reads, aligning the reads to a combination of the reference genome and a set of fusion-gene contigs based on the STAR-Fusion predictions. Reads mapping better to the fusion-gene contigs than the reference genome are identified and reported, fusions are re-scored/quantified, and fusion transcript allelic fractions are computed.
If either mode is invoked, STAR-Fusion will run FusionInspector and create a FusionInspector/ output subdirectory containing all relevant output files. See the FusionInspector Wiki for documentation on output files and loading results into IGV for visualization, or leveraging the html-based igv-reports.
An example html-report interactive fusion visualization is shown below:
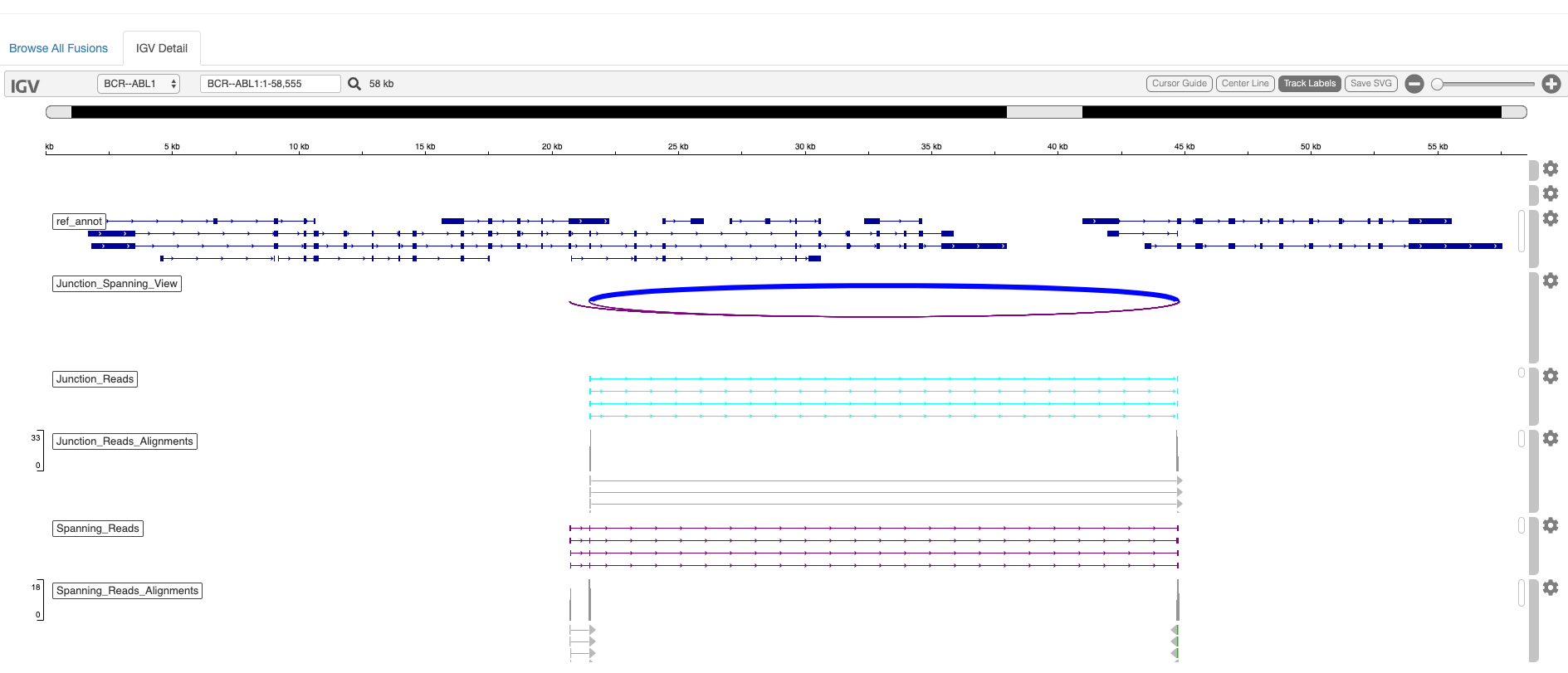
It is sometimes the case that fusion transcripts generate novel fusion proteins with altered functions. You can further explore the impact of the fusion event on coding regions by invoking the '--examine_coding_effect' parameter.
The coding effect results are appended as additional columns in the STAR-Fusion tab-delimited output file. An example set of columns include:
#FusionName BCR--ABL1
...
CDS_LEFT_ID ENST00000305877.8
CDS_LEFT_RANGE 1-2782
CDS_RIGHT_ID ENST00000318560.5
CDS_RIGHT_RANGE 80-3393
PROT_FUSION_TYPE INFRAME
FUSION_MODEL chr22|+|[0]23523148-23524426[0]|[1]23595986-23596167[2]|[0]23603137-23603241[2]|[0]23603542-23603727[2]|[0]23610595-23610702[2]|[0]23613719-23613779[0]|[1]23615268-23615320[2]|[0]23615821-23615961[2]|[0]23626164-23626285[1]|[2]23627220-23627388[2]|[0]23629346-23629465[2]|[0]23630284-23630359[0]|[1]23631704-23631808[0]|[1]23632526-23632600[0]<==>chr9|+|[1]133729451-133729624[0]|[1]133730188-133730483[2]|[0]133738150-133738422[2]|[0]133747516-133747600[0]|[1]133748247-133748424[1]|[2]133750255-133750439[0]|[1]133753802-133753954[0]|[1]133755455-133755544[0]|[1]133755887-133756051[0]|[1]133759356-133761070[2]
FUSION_CDS atggtggacccggtgggcttcgcggaggcgtggaaggcgcagttcccggactcagagcccccgcgcatggagctgcgctcagtgggcgacatcgagcaggagctggagcgctgcaaggcctccattcggcgcctggagcaggaggtgaaccaggagcgcttccgcatgatctacctgcagacgttgctggccaaggaaaagaagagctatgaccggcagcgatggggcttccggcgcgcggcgcaggcccccgacggcgcctccgagccccgagcgtccgcgtcgcgcccgcagccagcgcccgccgacggagccgacccgccgcccgccgaggagcccgaggcccggcccgacggcgagggttctccgggtaaggccaggcccgggaccgcccgcaggcccggggcagccgcgtcgggggaacgggacgaccggggaccccccgccagcgtggcggcgctcaggtccaacttcgagcggatccgcaagggccatggccagcccggggcggacgccgagaagcccttctacgtgaacgtcgagtttcaccacgagcgcggcctggtgaaggtcaacgacaaagaggtgtcggaccgcatcagctccctgggcagccaggccatgcagatggagcgcaaaaagtcccagcacggcgcgggctcgagcgtgggggatgcatccaggcccccttaccggggacgctcctcggagagcagctgcggcgtcgacggcgactacgaggacgccgagttgaacccccgcttcctgaaggacaacctgatcgacgccaatggcggtagcaggcccccttggccgcccctggagtaccagccctaccagagcatctacgtcgggggcatgatggaaggggagggcaagggcccgctcctgcgcagccagagcacctctgagcaggagaagcgccttacctggccccgcaggtcctactccccccggagttttgaggattgcggaggcggctataccccggactgcagctccaatgagaacctcacctccagcgaggaggacttctcctctggccagtccagccgcgtgtccccaagccccaccacctaccgcatgttccgggacaaaagccgctctccctcgcagaactcgcaacagtccttcgacagcagcagtccccccacgccgcagtgccataagcggcaccggcactgcccggttgtcgtgtccgaggccaccatcgtgggcgtccgcaagaccgggcagatctggcccaacgatggcgagggcgccttccatggagacgcagatggctcgttcggaacaccacctggatacggctgcgctgcagaccgggcagaggagcagcgccggcaccaagatgggctgccctacattgatgactcgccctcctcatcgccccacctcagcagcaagggcaggggcagccgggatgcgctggtctcgggagccctggagtccactaaagcgagtgagctggacttggaaaagggcttggagatgagaaaatgggtcctgtcgggaatcctggctagcgaggagacttacctgagccacctggaggcactgctgctgcccatgaagcctttgaaagccgctgccaccacctctcagccggtgctgacgagtcagcagatcgagaccatcttcttcaaagtgcctgagctctacgagatccacaaggagttctatgatgggctcttcccccgcgtgcagcagtggagccaccagcagcgggtgggcgacctcttccagaagctggccagccagctgggtgtgtaccgggccttcgtggacaactacggagttgccatggaaatggctgagaagtgctgtcaggccaatgctcagtttgcagaaatctccgagaacctgagagccagaagcaacaaagatgccaaggatccaacgaccaagaactctctggaaactctgctctacaagcctgtggaccgtgtgacgaggagcacgctggtcctccatgacttgctgaagcacactcctgccagccaccctgaccaccccttgctgcaggacgccctccgcatctcacagaacttcctgtccagcatcaatgaggagatcacaccccgacggcagtccatgacggtgaagaagggagagcaccggcagctgctgaaggacagcttcatggtggagctggtggagggggcccgcaagctgcgccacgtcttcctgttcaccgacctgcttctctgcaccaagctcaagaagcagagcggaggcaaaacgcagcagtatgactgcaaatggtacattccgctcacggatctcagcttccagatggtggatgaactggaggcagtgcccaacatccccctggtgcccgatgaggagctggacgctttgaagatcaagatctcccagatcaagaatgacatccagagagagaagagggcgaacaagggcagcaaggctacggagaggctgaagaagaagctgtcggagcaggagtcactgctgctgcttatgtctcccagcatggccttcagggtgcacagccgcaacggcaagagttacacgttcctgatctcctctgactatgagcgtgcagagtggagggagaacatccgggagcagcagaagaagtgtttcagaagcttctccctgacatccgtggagctgcagatgctgaccaactcgtgtgtgaaactccagactgtccacagcattccgctgaccatcaataaggaagatgatgagtctccggggctctatgggtttctgaatgtcatcgtccactcagccactggatttaagcagagttcaaAAGCCCTTCAGCGGCCAGTAGCATCTGACTTTGAGCCTCAGGGTCTGAGTGAAGCCGCTCGTTGGAACTCCAAGGAAAACCTTCTCGCTGGACCCAGTGAAAATGACCCCAACCTTTTCGTTGCACTGTATGATTTTGTGGCCAGTGGAGATAACACTCTAAGCATAACTAAAGGTGAAAAGCTCCGGGTCTTAGGCTATAATCACAATGGGGAATGGTGTGAAGCCCAAACCAAAAATGGCCAAGGCTGGGTCCCAAGCAACTACATCACGCCAGTCAACAGTCTGGAGAAACACTCCTGGTACCATGGGCCTGTGTCCCGCAATGCCGCTGAGTATCTGCTGAGCAGCGGGATCAATGGCAGCTTCTTGGTGCGTGAGAGTGAGAGCAGTCCTGGCCAGAGGTCCATCTCGCTGAGATACGAAGGGAGGGTGTACCATTACAGGATCAACACTGCTTCTGATGGCAAGCTCTACGTCTCCTCCGAGAGCCGCTTCAACACCCTGGCCGAGTTGGTTCATCATCATTCAACGGTGGCCGACGGGCTCATCACCACGCTCCATTATCCAGCCCCAAAGCGCAACAAGCCCACTGTCTATGGTGTGTCCCCCAACTACGACAAGTGGGAGATGGAACGCACGGACATCACCATGAAGCACAAGCTGGGCGGGGGCCAGTACGGGGAGGTGTACGAGGGCGTGTGGAAGAAATACAGCCTGACGGTGGCCGTGAAGACCTTGAAGGAGGACACCATGGAGGTGGAAGAGTTCTTGAAAGAAGCTGCAGTCATGAAAGAGATCAAACACCCTAACCTGGTGCAGCTCCTTGGGGTCTGCACCCGGGAGCCCCCGTTCTATATCATCACTGAGTTCATGACCTACGGGAACCTCCTGGACTACCTGAGGGAGTGCAACCGGCAGGAGGTGAACGCCGTGGTGCTGCTGTACATGGCCACTCAGATCTCGTCAGCCATGGAGTACCTGGAGAAGAAAAACTTCATCCACAGAGATCTTGCTGCCCGAAACTGCCTGGTAGGGGAGAACCACTTGGTGAAGGTAGCTGATTTTGGCCTGAGCAGGTTGATGACAGGGGACACCTACACAGCCCATGCTGGAGCCAAGTTCCCCATCAAATGGACTGCACCCGAGAGCCTGGCCTACAACAAGTTCTCCATCAAGTCCGACGTCTGGGCATTTGGAGTATTGCTTTGGGAAATTGCTACCTATGGCATGTCCCCTTACCCGGGAATTGACCTGTCCCAGGTGTATGAGCTGCTAGAGAAGGACTACCGCATGGAGCGCCCAGAAGGCTGCCCAGAGAAGGTCTATGAACTCATGCGAGCATGTTGGCAGTGGAATCCCTCTGACCGGCCCTCCTTTGCTGAAATCCACCAAGCCTTTGAAACAATGTTCCAGGAATCCAGTATCTCAGACGAAGTGGAAAAGGAGCTGGGGAAACAAGGCGTCCGTGGGGCTGTGAGTACCTTGCTGCAGGCCCCAGAGCTGCCCACCAAGACGAGGACCTCCAGGAGAGCTGCAGAGCACAGAGACACCACTGACGTGCCTGAGATGCCTCACTCCAAGGGCCAGGGAGAGAGCGATCCTCTGGACCATGAGCCTGCCGTGTCTCCATTGCTCCCTCGAAAAGAGCGAGGTCCCCCGGAGGGCGGCCTGAATGAAGATGAGCGCCTTCTCCCCAAAGACAAAAAGACCAACTTGTTCAGCGCCTTGATCAAGAAGAAGAAGAAGACAGCCCCAACCCCTCCCAAACGCAGCAGCTCCTTCCGGGAGATGGACGGCCAGCCGGAGCGCAGAGGGGCCGGCGAGGAAGAGGGCCGAGACATCAGCAACGGGGCACTGGCTTTCACCCCCTTGGACACAGCTGACCCAGCCAAGTCCCCAAAGCCCAGCAATGGGGCTGGGGTCCCCAATGGAGCCCTCCGGGAGTCCGGGGGCTCAGGCTTCCGGTCTCCCCACCTGTGGAAGAAGTCCAGCACGCTGACCAGCAGCCGCCTAGCCACCGGCGAGGAGGAGGGCGGTGGCAGCTCCAGCAAGCGCTTCCTGCGCTCTTGCTCCGCCTCCTGCGTTCCCCATGGGGCCAAGGACACGGAGTGGAGGTCAGTCACGCTGCCTCGGGACTTGCAGTCCACGGGAAGACAGTTTGACTCGTCCACATTTGGAGGGCACAAAAGTGAGAAGCCGGCTCTGCCTCGGAAGAGGGCAGGGGAGAACAGGTCTGACCAGGTGACCCGAGGCACAGTAACGCCTCCCCCCAGGCTGGTGAAAAAGAATGAGGAAGCTGCTGATGAGGTCTTCAAAGACATCATGGAGTCCAGCCCGGGCTCCAGCCCGCCCAACCTGACTCCAAAACCCCTCCGGCGGCAGGTCACCGTGGCCCCTGCCTCGGGCCTCCCCCACAAGGAAGAAGCTGGAAAGGGCAGTGCCTTAGGGACCCCTGCTGCAGCTGAGCCAGTGACCCCCACCAGCAAAGCAGGCTCAGGTGCACCAGGGGGCACCAGCAAGGGCCCCGCCGAGGAGTCCAGAGTGAGGAGGCACAAGCACTCCTCTGAGTCGCCAGGGAGGGACAAGGGGAAATTGTCCAGGCTCAAACCTGCCCCGCCGCCCCCACCAGCAGCCTCTGCAGGGAAGGCTGGAGGAAAGCCCTCGCAGAGCCCGAGCCAGGAGGCGGCCGGGGAGGCAGTCCTGGGCGCAAAGACAAAAGCCACGAGTCTGGTTGATGCTGTGAACAGTGACGCTGCCAAGCCCAGCCAGCCGGGAGAGGGCCTCAAAAAGCCCGTGCTCCCGGCCACTCCAAAGCCACAGTCCGCCAAGCCGTCGGGGACCCCCATCAGCCCAGCCCCCGTTCCCTCCACGTTGCCATCAGCATCCTCGGCCCTGGCAGGGGACCAGCCGTCTTCCACCGCCTTCATCCCTCTCATATCAACCCGAGTGTCTCTTCGGAAAACCCGCCAGCCTCCAGAGCGGATCGCCAGCGGCGCCATCACCAAGGGCGTGGTCCTGGACAGCACCGAGGCGCTGTGCCTCGCCATCTCTAGGAACTCCGAGCAGATGGCCAGCCACAGCGCAGTGCTGGAGGCCGGCAAAAACCTCTACACGTTCTGCGTGAGCTATGTGGATTCCATCCAGCAAATGAGGAACAAGTTTGCCTTCCGAGAGGCCATCAACAAACTGGAGAATAATCTCCGGGAGCTTCAGATCTGCCCGGCGACAGCAGGCAGTGGTCCAGCGGCCACTCAGGACTTCAGCAAGCTCCTCAGTTCGGTGAAGGAAATCAGTGACATAGTGCAGAGGTAG
FUSION_TRANSL MVDPVGFAEAWKAQFPDSEPPRMELRSVGDIEQELERCKASIRRLEQEVNQERFRMIYLQTLLAKEKKSYDRQRWGFRRAAQAPDGASEPRASASRPQPAPADGADPPPAEEPEARPDGEGSPGKARPGTARRPGAAASGERDDRGPPASVAALRSNFERIRKGHGQPGADAEKPFYVNVEFHHERGLVKVNDKEVSDRISSLGSQAMQMERKKSQHGAGSSVGDASRPPYRGRSSESSCGVDGDYEDAELNPRFLKDNLIDANGGSRPPWPPLEYQPYQSIYVGGMMEGEGKGPLLRSQSTSEQEKRLTWPRRSYSPRSFEDCGGGYTPDCSSNENLTSSEEDFSSGQSSRVSPSPTTYRMFRDKSRSPSQNSQQSFDSSSPPTPQCHKRHRHCPVVVSEATIVGVRKTGQIWPNDGEGAFHGDADGSFGTPPGYGCAADRAEEQRRHQDGLPYIDDSPSSSPHLSSKGRGSRDALVSGALESTKASELDLEKGLEMRKWVLSGILASEETYLSHLEALLLPMKPLKAAATTSQPVLTSQQIETIFFKVPELYEIHKEFYDGLFPRVQQWSHQQRVGDLFQKLASQLGVYRAFVDNYGVAMEMAEKCCQANAQFAEISENLRARSNKDAKDPTTKNSLETLLYKPVDRVTRSTLVLHDLLKHTPASHPDHPLLQDALRISQNFLSSINEEITPRRQSMTVKKGEHRQLLKDSFMVELVEGARKLRHVFLFTDLLLCTKLKKQSGGKTQQYDCKWYIPLTDLSFQMVDELEAVPNIPLVPDEELDALKIKISQIKNDIQREKRANKGSKATERLKKKLSEQESLLLLMSPSMAFRVHSRNGKSYTFLISSDYERAEWRENIREQQKKCFRSFSLTSVELQMLTNSCVKLQTVHSIPLTINKEDDESPGLYGFLNVIVHSATGFKQSSKALQRPVASDFEPQGLSEAARWNSKENLLAGPSENDPNLFVALYDFVASGDNTLSITKGEKLRVLGYNHNGEWCEAQTKNGQGWVPSNYITPVNSLEKHSWYHGPVSRNAAEYLLSSGINGSFLVRESESSPGQRSISLRYEGRVYHYRINTASDGKLYVSSESRFNTLAELVHHHSTVADGLITTLHYPAPKRNKPTVYGVSPNYDKWEMERTDITMKHKLGGGQYGEVYEGVWKKYSLTVAVKTLKEDTMEVEEFLKEAAVMKEIKHPNLVQLLGVCTREPPFYIITEFMTYGNLLDYLRECNRQEVNAVVLLYMATQISSAMEYLEKKNFIHRDLAARNCLVGENHLVKVADFGLSRLMTGDTYTAHAGAKFPIKWTAPESLAYNKFSIKSDVWAFGVLLWEIATYGMSPYPGIDLSQVYELLEKDYRMERPEGCPEKVYELMRACWQWNPSDRPSFAEIHQAFETMFQESSISDEVEKELGKQGVRGAVSTLLQAPELPTKTRTSRRAAEHRDTTDVPEMPHSKGQGESDPLDHEPAVSPLLPRKERGPPEGGLNEDERLLPKDKKTNLFSALIKKKKKTAPTPPKRSSSFREMDGQPERRGAGEEEGRDISNGALAFTPLDTADPAKSPKPSNGAGVPNGALRESGGSGFRSPHLWKKSSTLTSSRLATGEEEGGGSSSKRFLRSCSASCVPHGAKDTEWRSVTLPRDLQSTGRQFDSSTFGGHKSEKPALPRKRAGENRSDQVTRGTVTPPPRLVKKNEEAADEVFKDIMESSPGSSPPNLTPKPLRRQVTVAPASGLPHKEEAGKGSALGTPAAAEPVTPTSKAGSGAPGGTSKGPAEESRVRRHKHSSESPGRDKGKLSRLKPAPPPPPAASAGKAGGKPSQSPSQEAAGEAVLGAKTKATSLVDAVNSDAAKPSQPGEGLKKPVLPATPKPQSAKPSGTPISPAPVPSTLPSASSALAGDQPSSTAFIPLISTRVSLRKTRQPPERIASGAITKGVVLDSTEALCLAISRNSEQMASHSAVLEAGKNLYTFCVSYVDSIQQMRNKFAFREAINKLENNLRELQICPATAGSGPAATQDFSKLLSSVKEISDIVQR*
PFAM_LEFT Bcr-Abl_Oligo|3-75|3.6e-43^RhoGEF|503-689|1.1e-37^IQ_SEC7_PH|724-769|4.7e-06^PH_5|727-865|6.6e-05^PH|740-864|5.3e-06^Bcr-Abl_Oligo|781-802|0.45^C2|913-1007|2.2e-09^PH|919-973|0.56^RhoGAP|1068-1218|7.9e-52
PFAM_RIGHT SH3_1-PARTIAL|~80-113|1.6e-15^SH3_9-PARTIAL|~80-117|1.3e-11^SH3_2-PARTIAL|~80-118|1.9e-10^SH3_3|82-116|9.2e-08^SH3_6|83-112|0.00021^SH2|127-202|1.4e-26^Pkinase_Tyr|242-492|3.3e-102^Pkinase|244-491|2.1e-51^Haspin_kinase|307-388|9.1e-07^F_actin_bind|1027-1130|2.2e-33
The 'FusionModel' describes the gene structure of the reconstructed fusion transcript model given the breakpoint and preferentially selecting in-frame coding structures among multiple isoforms. The format is "chr|strand|[exon_start_codon_phase]|lend-rend[exon_end_codon_phase]...<==> info for connecting partner in the same format".
The PFAM information corresponds to the positions of the domains in the putative fusion protein, based on domain positions within the original protein annotations. A ~ symbol indicates that the breakpoint fractures the original domain annotation.
Note, fusion annotation and examination of coding effects are additionally performed on FusionInspector outputs if '--FusionInspector inspect|validate' is invoked in your STAR-Fusion run.
A utility script is provided to append the breakpoint adjacent sequences that could be used for designing primers for PCR based validation:
STAR-Fusion/util/append_fusion_brkpt_adjacent_sequence.pl star-fusion.fusion_predictions.abridged.tsv $CTAT_GENOME_LIB/
A column 'FusionSeq' is added providing the breakpoint-adjacent sequences, in upper/lowercase for alternate sides of the breakpoint.
0 THRA--AC090627.1
1 30
2 93
3 30.00
4 91.96
5 ONLY_REF_SPLICE
6 THRA^ENSG00000126351.11
7 chr17:40086853:+
8 AC090627.1^ENSG00000235300.4
9 chr17:48294347:+
10 YES_LDAS
11 81036.5448
12 GT
13 1.8892
14 AG
15 1.9656
16 ["INTRACHROMOSOMAL[chr17:8.20Mb]"]
17 CCATCACCCGTGTGGTGGACTTTGCCAAAAAACTGCCCATGTTCTCCGAGcaatttcgagtgcaagtgccacagtgtcagctaaagaaacacaactgcgg
The reconstructed transcripts described above by the 'fusion coding effect' module are based on the reference annotations and the reference genome sequence. If you're interested in doing de novo reconstruction of fusion transcripts based on the actual fusion-supporting RNA-Seq reads, capturing any additional variant information or novel sequence features that may be evidence among the reads, you can include de novo reconstruction by invoking the STAR-Fusion '--denovo_reconstruct' parameter. This requires that you include the '--FusionInspector inspect|validate' setting. Based on the STAR-Fusion predicted fusions.
Trinity is used to assemble reads aligned to fusion contigs constructed by FusionInspector. If FusionInspector 'inspect' mode is invoked, then only the fusion-evidence reads are de novo assembled. If FusionInspector 'validate' mode is selected, then all reads aligned to the fusion gene contigs are assembled. Trinity contigs identified as fusion transcripts are captured and reported. The de novo reconstructed fusion transcripts are provided as a FASTA file '${star_fusion_output_directory}/FusionInspector/finspector.gmap_trinity_GG.fusions.fasta', and the transcript accessions are reported in the FusionInspector tab-delimited summary output file.
An example Trinity-reconstructed fusion transcript is:
>TRINITY_GG_12_c0_g1_i1 VAPB--IKZF3:1396-28254
CGGTGTCTGGACCAAGGGGCGCAGGGCTTCGGCGCCAAGATAGCTGATGGCGTTATTGATGGCTTGGTCCATCATGCGGG
TCTGTATGAGCTCACTCTCTTTCTCATACATGTAACTTGAATTATAGTTGACATCAAAGCAGTGGCGCTTCTCACCAATG
AATTTCTGAGGCATTGAGCTTTTTCGTTTTGCCACATTGCTTGCTAATCTGTCCAGTACGAGAGCTCTTTCACTTCCCAT
CTCTGCTTTGATGTGTCTTGCCTCCGCACTTGCTCGGAATTTGAGCTCGTGCTGCGGCTCGAGGCTCAGGACCTGCTCCA
CCTTCGCCATGTTCCTTAGCGGCGGAGCACCTTTGGCGGGGAGACCCCTGAGAGGTCACCGGGGCGGGAAGCGTTAATGC
TGCGCCCGCTTTAAGTTTTACAAAAAGGCGGGGACCGGTCGGGGCACGGGCGGGGGTCCTCTACCG
See our FusionInspector wiki for more details.
In the included testing/ directory, you'll find a small sample of fastq reads from a tumor sample. Find fusions using the resource set like so:
cd testing/
../STAR-Fusion --left_fq reads_1.fq.gz --right_fq reads_2.fq.gz \
-O star_fusion_outdir \
--genome_lib_dir /path/to/your/CTAT_resource_lib \
--verbose_level 2

We provide a Docker image that contains all software pre-installed for running STAR, STAR-Fusion, and FusionInspector, and it's available here: https://hub.docker.com/r/trinityctat/starfusion/
If you have docker installed, you can pull the image like so:
docker pull trinityctat/starfusion
STAR-Fusion could be run like so via Docker, for example, running within the '${STAR_FUSION_HOME}/Docker' folder, where ${STAR_FUSION_HOME} is your base installation directory for the STAR-Fusion software.
# and now running STAR-Fusion & FusionInspector 'inspect' & Trinity de-novo reconstruction via Docker,
# below we assume you have your reads_1.fq.gz and reads_2.fq.gz in your current working directory
# and also have the ctat_genome_lib_build_dir in your current directory.
docker run -v `pwd`:/data --rm trinityctat/starfusion \
STAR-Fusion \
--left_fq /data/reads_1.fq.gz \
--right_fq /data/reads_2.fq.gz \
--genome_lib_dir /data/ctat_genome_lib_build_dir \
-O /data/StarFusionOut \
--FusionInspector validate \
--examine_coding_effect \
--denovo_reconstruct

Singularity is easier and safer to use than Docker, and is our preferred method for running STAR-Fusion. All modern releases of STAR-Fusion have a Singularity image (.simg) offered for download from our STAR-Fusion Singularity Image Release Site. If you have Singularity installed and the .simg file downloaded, you can run STAR-Fusion like so:
singularity exec -e -B `pwd` -B /path/to/ctat_genome_lib_build_dir \
star-fusion-v$version.simg \
STAR-Fusion \
--left_fq reads_1.fq.gz \
--right_fq reads_2.fq.gz \
--genome_lib_dir /path/to/ctat_genome_lib_build_dir \
-O StarFusionOut \
--FusionInspector validate \
--examine_coding_effect \
--denovo_reconstruct
Terra is a cloud-native platform for biomedical researchers to access data, run analysis tools, and collaborate.
Please see running STAR-Fusion on Terra for instructions.
Questions, comments, etc?
Visit our STAR-fusion Google group https://groups.google.com/forum/#!forum/star-fusion
This effort was largely inspired by earlier work done by Nicolas Stransky in the landmark publication "The landscape of kinase fusions in cancer" by Stransky et al., Nat Commun 2014, in addition to very nice work done by Daniel Nicorici with his FusionCatcher software.
STAR-Fusion is contributed by Brian Haas (Broad Institute), in collaboration with Alex Dobin (Cold Spring Harbor Laboratory). STAR-Fusion is several components being developed as part of the Trinity Cancer Transcriptomics Toolkit.
STAR-Fusion should be cited as:
Accuracy assessment of fusion transcript detection via read-mapping and de novo fusion transcript assembly-based methods. Haas, Brian J.; Dobin, Alexander; Li, Bo; Stransky, Nicolas; Pochet, Nathalie; Regev, Aviv; Genome Biology; 2019 https://genomebiology.biomedcentral.com/articles/10.1186/s13059-019-1842-9
Our earlier pre-print (unpublished):
STAR-Fusion: Fast and Accurate Fusion Transcript Detection from RNA-Seq Brian Haas, Alexander Dobin, Nicolas Stransky, Bo Li, Xiao Yang, Timothy Tickle, Asma Bankapur, Carrie Ganote, Thomas Doak, Natalie Pochet, Jing Sun, Catherine Wu, Thomas Gingeras, Aviv Regev bioRxiv 120295; doi: https://doi.org/10.1101/120295 https://www.biorxiv.org/content/10.1101/120295v1
Data sets leveraged for the above paper are available on Zenodo as:
- simulated 50 PE fusion reads and template fusion target sequences
- simulated 101 PE fusion reads
- cancer cell line reads or names of reads in the case of CCLE. For CCLE sequence data, visit the CCLE site.
STAR-Fusion is supported as part of the Trinity CTAT Project, funded by the National Cancer Institute Informatics Technology for Cancer Research
- STAR-Fusion Wiki Home
- Installing STAR-Fusion
- Software Compatibility Matrix
- Computing/Hardware Requirements
-
Running STAR-Fusion
- Running from FASTQ files
- Kickstart mode
- Run using Docker or Singularity
- Run using Terra
- Fusion Outputs
- Inspection and Validation
- Extract breakpoint-adjacent sequences
- Examine Fusion Effect on Coding Regions
- De novo reconstruct fusion transcripts
- Single cell RNAseq fusion detection
- Tutorial with example data and execution
- FAQ
- Contact us