Genomics Status is a Tornado web app for visualizing information and statistics regarding SciLifeLab Genomics platform operations.
Status interfaces with StatusDB; the CouchDB database instance we're using at SciLifeLab to store metadata in various forms. Document specifications for StatusDB are available in the internal wiki. Documentation about CouchDB can be found here.
NOTE: These steps assume that:
- you're either running a python virtualenv or you do have root permissions.
- you're running python version 3.6 or later (dicts are ordered by default)
- you have access to both StatusDB and Genologics LIMS
1 - Clone the repository with the --recursive option (this will also download nvd3 library):
git clone --recursive https://github.com/NationalGenomicsInfrastructure/genomics-status.git
2 - Install the package (the pip install -r requirements_dev.txt can be skipped on a production server)
cd status
pip install -r requirements.txt
pip install -r requirements_dev.txt
python setup.py install
3 - You'll need to install genologics package separately:
pip install git+https://github.com/SciLifeLab/genologics.git
4 - For running, it requires a settings.yaml file which points to the CouchDB server to use, and which port to
serve the web app to. You will also need a .genologicsrc file with the API credentials for our Genologics LIMS. The files should look like these:
<status_dir>/run_dir/settings.yaml:
couch_server: http://<username>:<password>@ngi-statusdb-dev.scilifelab.se:5984
username: <tools_username> # same as input above
password: <tools_password> # same as input above
port: 9761
redirect_uri: http://localhost:9761/login
# This can be left like this as long as --testing_mode is used
google_oauth:
key_old: write
key: anything
secret: here
zendesk:
url: any
username: thing
token: goes
sftp:
login: will
password: code
contact_person: [email protected]
instruments:
HiSeq:
INSTRUMENT_ID: INSTRUMENT_NAME
MiSeq:
INSTRUMENT_ID: INSTRUMENT_NAME
password_seed: not_needed_for_development
# You Trello API credentials, you need a board named "Suggestion Box"
trello:
api_key: <trello_api_key>
api_secret: <trello_api_secret>
token: <trello_token>
uppmax_projects
- proj_id1
- proj_id2~/.genologicsrc:
[genologics]
BASEURI=https://ngi-lims-stage.scilifelab.se:8443
USERNAME=<lims_api_username>
PASSWORD=<lims_api_password>5 - Run the tornado app from run_dir :
cd run_dir
python ../status_app.py --testing_mode
--testing_mode will skip the google authentication, which is convenient for testing
The status web app both provides the HTML web interface, and a RESTful api for accessing the data being visualized on the various pages.
If you've used the settings.yaml template above, you should now be able to access the site at http://localhost:9761/ or http://localhost:9761/login
The bootstrap css file is customised to use a smaller font size. To replicate this, use the following snippet:
conda create -n nodejs
conda activate nodejs
conda install nodejs
npm install sass -g
npm install bootstrap@next -g
cd run_dir/static/scss
Then modify the custom.scss file, especially the hard-coded path according to your needs, and run the compilation:
sass custom.scss custom-bootstrap-5-alpha.css
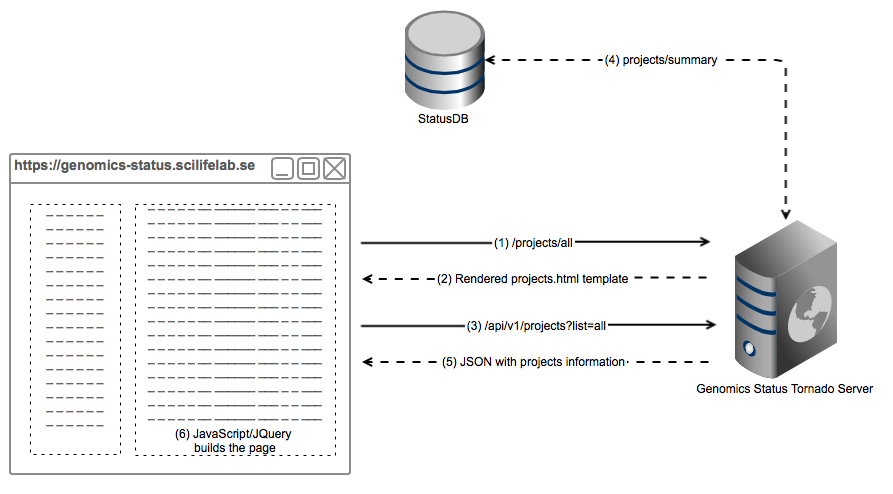
This pictures illustrates the architecture of how Genomics Status is built with a real example, a request to https://genomics-status.scilifelab.se/projects/all. It is simplified for the sake of comprehension, in reality there are a few more API calls.
- The web browser (a human, actually) requests the page
/projects/all. The browser sends the request to Tornado, which has assigned theProjectsHandlerto this call. - Tornado returns a rendered template with all the parameters needes to build the projects page, i.e username, projects to list, etc.
- Within the template, in order to build the project list, it performas a JavaScript (JQuery) call to GenStat API.
- Tornado queries StatusDB information about the projects and parses it correctly.
- A JSON document is returned to the web browser
- Which uses it to build the project list client-side.
This design aims to decouple design and backend, as well as avoid making calls to the database from the web browser.
It also facilitates the reusability of the API for other possible applications.
Tornado is a Python web framework and asynchronous networking library. Genomics Status is based on Tornado.
A very basic tornado web app, but enough to get the idea, would be something like this:
import tornado.ioloop
import tornado.web
class MainHandler(tornado.web.RequestHandler):
def get(self):
self.write("Hello, world")
application = tornado.web.Application([
(r"/", MainHandler),
])
if __name__ == "__main__":
application.listen(8888)
tornado.ioloop.IOLoop.instance().start()Basically, you have to define a handlaer for each URL you want your application to serve. In this case, we define just one handler for the URI '/'. This will just print a "Hello, World" page.
Handlers that inherit from tornado.web.RequestHandler should implement at least one of the HTTP basic operations, i.e GET, POST or PUT.
Tornado templates are a good way to generate dynamic pages server side. The advantage of templates is that you can embeed python code in them. The official documentation is good enough to learn how they work.
On Genomics Status, templates are located in status/run_dir/design