-
Notifications
You must be signed in to change notification settings - Fork 0
Commit
This commit does not belong to any branch on this repository, and may belong to a fork outside of the repository.
- Loading branch information
Showing
6 changed files
with
522 additions
and
1 deletion.
There are no files selected for viewing
14 changes: 14 additions & 0 deletions
14
_freeze/posts/2023/2023-12-10-qPCR---Repeat-CAct-Primer-Test/index/execute-results/html.json
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| Original file line number | Diff line number | Diff line change |
|---|---|---|
| @@ -0,0 +1,14 @@ | ||
| { | ||
| "hash": "44619ea66eaca52463eb30794cdcf8fd", | ||
| "result": { | ||
| "markdown": "---\nauthor: Sam White\ntoc-title: Contents\ntoc-depth: 5\ntoc-location: left\ntitle: qPCR - Repeat CAct Primer Test\ndate: '2024-12-10'\ndraft: true\nengine: knitr\ncategories: \n - 2023\n - CFX Connect\n - qPCR\n - CAct\n---\n\n# Intro\n\nDue to odd results from [earlier today](https://jpglabhome.github.io/SymbID_protbleach/posts/2023-12-10-qPCR---Repeat-CAct-Primer-Test/) (Notebook), I repeated this qPCR to see if I could get better replication and consistent melt curves across samples.\n\n## qPCRs\nAll qPCRs were run in triplicate for each dilution (1000pg, 100pg, 10pg), with SsoAdvanced Universal SYBR Green Supermix in 20uL reactions. The CAct primers were utilized with an initial “working stock” concentration of 10uM and at a final concentration of 0.25uM in each qPCR reaction.\n\n## qPCR calculations\n\n- [20231210-qPCR_calcs](https://docs.google.com/spreadsheets/d/1dnuJzmNqAzvgVht9TEDm5e1sXGLLLcDxT8YvYFmLQbg/edit?usp=sharing) (Google Sheet)\n\n# Results\n\n## Output files\n\n### Raw qPCR data (`.pcrd`; Requires CFX Maestro):\n\n- [`sam_2023-12-10_09-26-24_Connect.pcrd`](../../../data/qPCR/testing/raw/sam_2023-12-10_09-26-24_Connect.pcrd)\n\n - \"Analysis Mode\" was set to \"Target\" (\"Fluorophore\" is the default.)\n\n - Baseline threshold was set to \"Regression\"\n\n### qPCR data (CSV):\n\n- [`sam_2023-12-10_09-26-24_Connect-Quantification-Cq_Results.csv`](../../../data/qPCR/testing/exported/sam_2023-12-10_09-26-24_Connect-Quantification-Cq_Results.csv)\n\n### qPCR Report (PDF):\n\n- [`sam_2023-12-10_07-50-37_Connect.pdf`](../../../data/qPCR/testing/cfx_reports/sam_2023-12-10_09-26-24_Connect.pdf)\n\n::: {layout-nrow=\"1\"}\n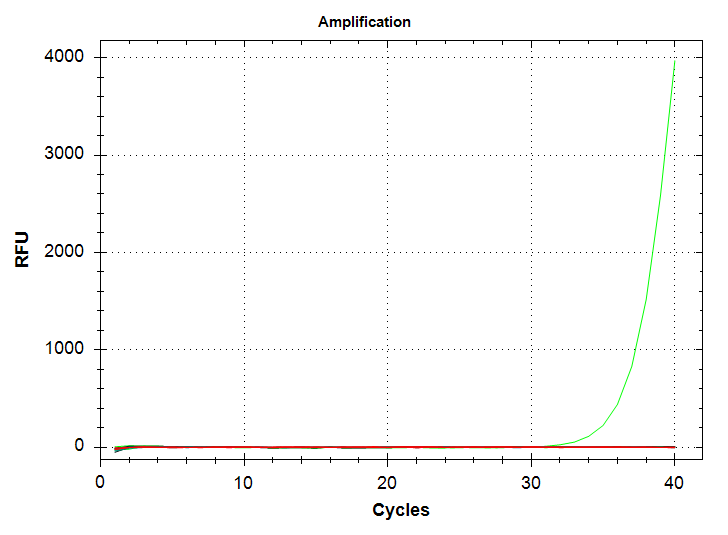{fig-alt=\"CAct amplification plot. Green line is 1000pg input DNA and corresponds to a Cq value of ~37.\"}\n\n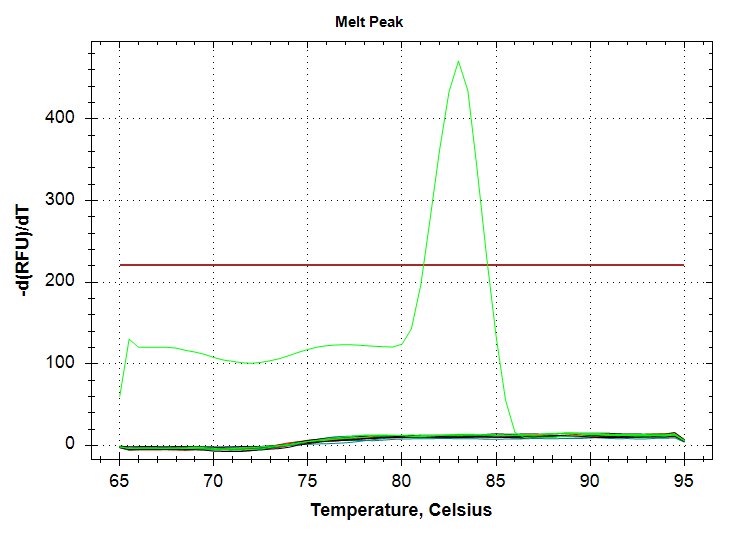{fig-alt=\"CAct melt plot. Green peak is 1000ng input DNA.\"}\n:::\n\nWell, the results are similar to the previous run, in that there's a single 1000pg replicate which amplifies at a Cq of ~37, while we no longer see amplification in any 100pg replicates. Admittedly, the latter was expected, based on the [different melting temp of the 100pg product seen previously](https://jpglabhome.github.io/SymbID_protbleach/posts/2023-12-10-qPCR---Repeat-CAct-Primer-Test/); i.e. the previous amplification of 100pg DNA wasn't \"real.\"\n\nHowever, what was not expected was my continued inability to produce proper technical replicates. I'm wondering if the sensitivity of this primer is low enough that it's causing a large (i.e. ~3 Cq) swing in Cqs between replicates. I'll repeat this using the 10,000pg (10ng) stock to see if this improves replication...", | ||
| "supporting": [], | ||
| "filters": [ | ||
| "rmarkdown/pagebreak.lua" | ||
| ], | ||
| "includes": {}, | ||
| "engineDependencies": {}, | ||
| "preserve": {}, | ||
| "postProcess": true | ||
| } | ||
| } |
Binary file added
BIN
+169 KB
docs/data/qPCR/testing/cfx_reports/sam_2023-12-10_09-26-24_Connect.pdf
Binary file not shown.
13 changes: 13 additions & 0 deletions
13
.../data/qPCR/testing/exported/sam_2023-12-10_09-26-24_Connect-Quantification-Cq_Results.csv
This file contains bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| Original file line number | Diff line number | Diff line change |
|---|---|---|
| @@ -0,0 +1,13 @@ | ||
| ,Well,Fluor,Target,Content,Sample,Biological Set Name,Cq,Cq Mean,Cq Std. Dev,Starting Quantity (SQ),Log Starting Quantity,SQ Mean,SQ Std. Dev,Set Point,Well Note | ||
| ,A01,SYBR,CAct,Unkn-1,1000pg,,36.6197981615186,36.6197981615186,0,NaN,NaN,NaN,0,60, | ||
| ,A02,SYBR,CAct,Unkn-1,1000pg,,NaN,0,0,NaN,NaN,0,0,60, | ||
| ,A03,SYBR,CAct,Unkn-1,1000pg,,NaN,0,0,NaN,NaN,0,0,60, | ||
| ,A04,SYBR,CAct,Unkn-2,100pg,,NaN,0,0,NaN,NaN,0,0,60, | ||
| ,A05,SYBR,CAct,Unkn-2,100pg,,NaN,0,0,NaN,NaN,0,0,60, | ||
| ,A06,SYBR,CAct,Unkn-2,100pg,,NaN,0,0,NaN,NaN,0,0,60, | ||
| ,A07,SYBR,CAct,Unkn-3,10pg,,NaN,0,0,NaN,NaN,0,0,60, | ||
| ,A08,SYBR,CAct,Unkn-3,10pg,,NaN,0,0,NaN,NaN,0,0,60, | ||
| ,B01,SYBR,CAct,Unkn-3,10pg,,NaN,0,0,NaN,NaN,0,0,60, | ||
| ,B02,SYBR,CAct,NTC-1,,,NaN,0,0,NaN,NaN,0,0,60, | ||
| ,B03,SYBR,CAct,NTC-1,,,NaN,0,0,NaN,NaN,0,0,60, | ||
| ,B04,SYBR,CAct,NTC-1,,,NaN,0,0,NaN,NaN,0,0,60, |
Binary file not shown.
Oops, something went wrong.