Visualization of RNA secondary structure using a template visualization (currently Comparative RNA Web (CRW) Site postscripts and VARNA SVG formats are supported). The first version of Traveler was developed by Richard Elias (and still the abosulte majority of the code is his work) and the original repository should be accessible at https://github.com/rikiel/traveler .
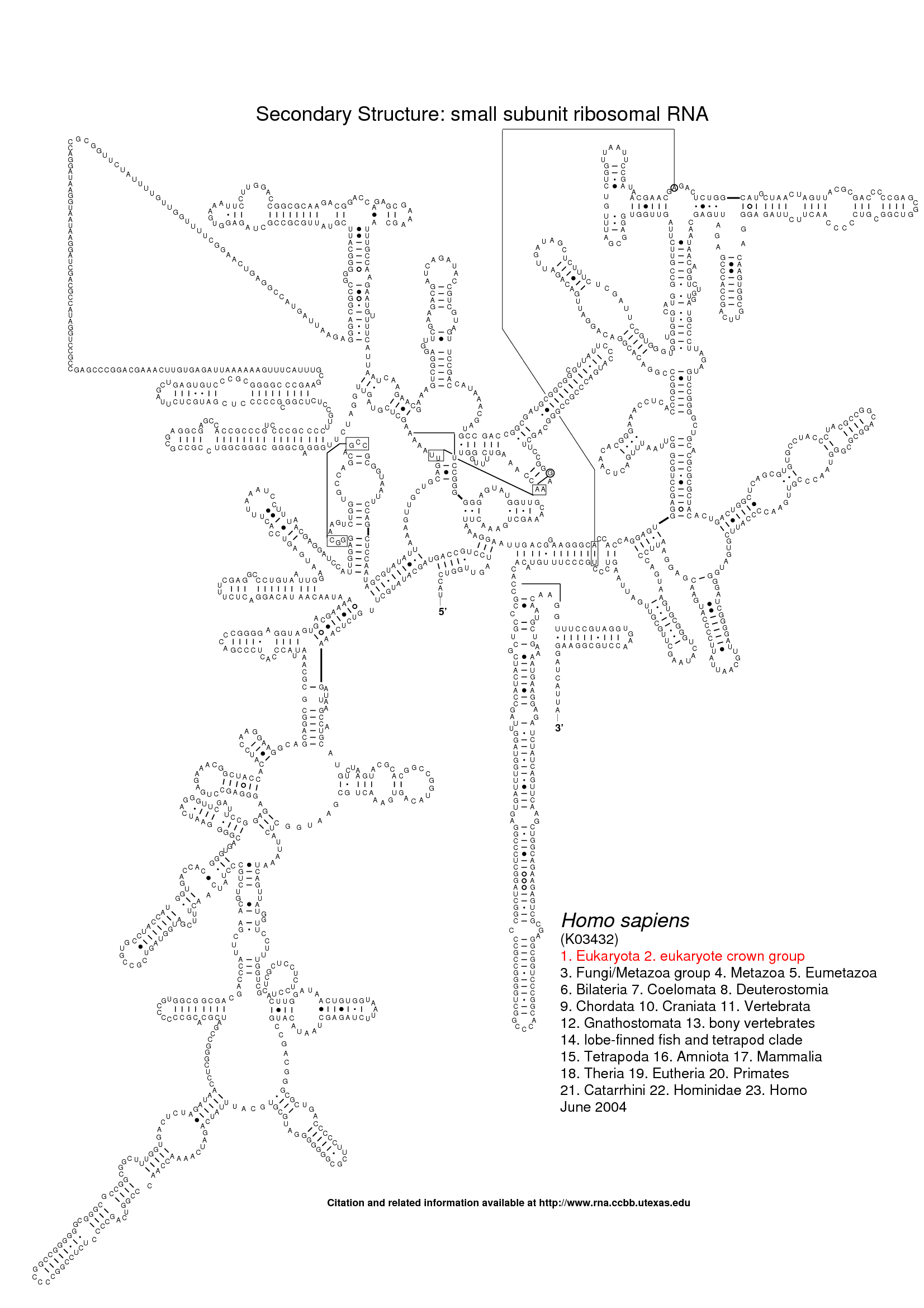
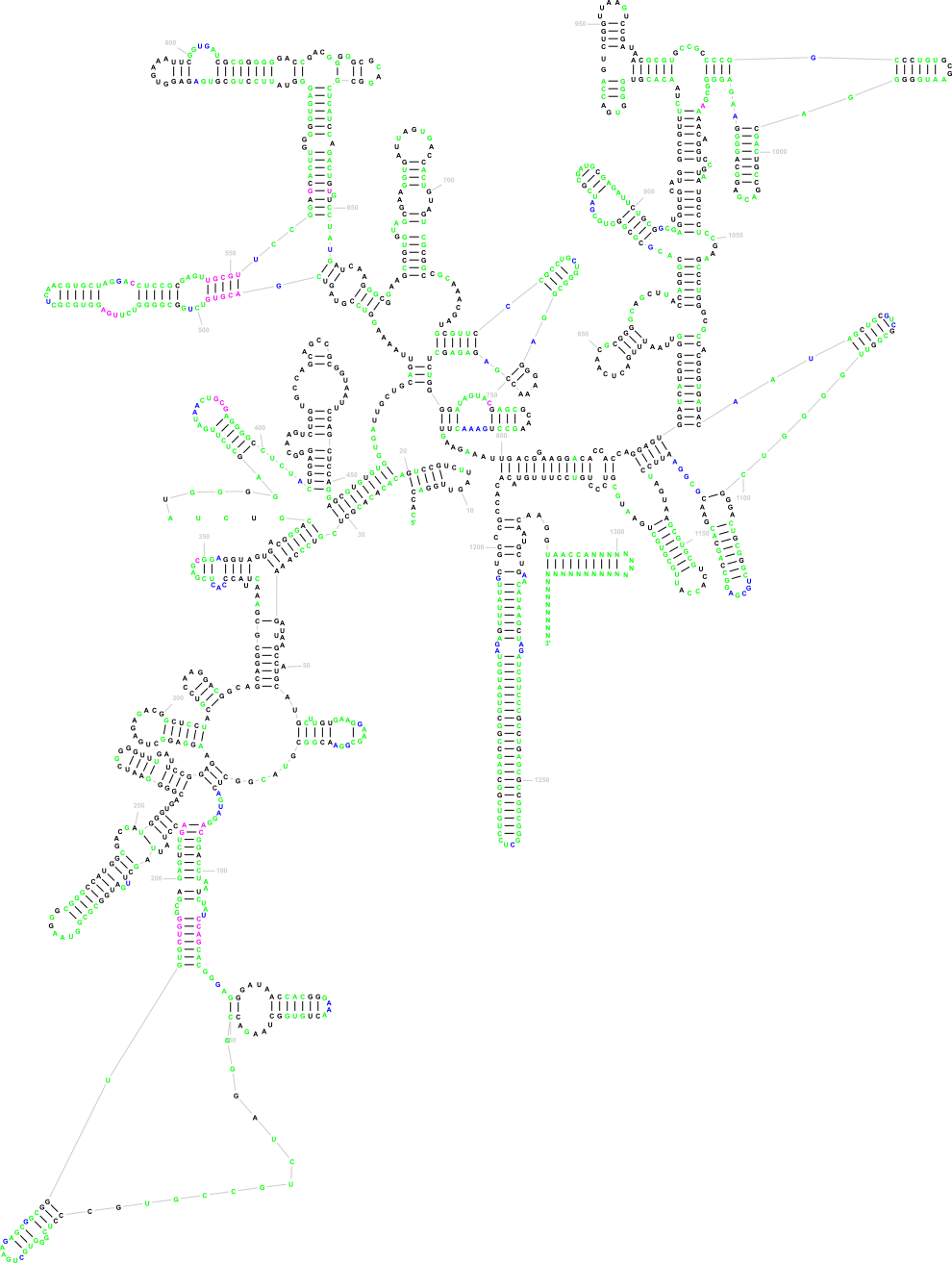
The following three images show what Traveler is good for. The first image is the visualization of 18S human rRNA from CRW, follows the correct visualization of Amblyospora bracteata's (AB) 18s rRNA (as downloaded from CRW) and the third image shows how Traveler visualizes the AB's 18s when given its secondary structure together with human rRNA secondary structure and visualization in postscript as a template.
- gcc with support of c++11
Use git clone https://github.com/davidhoksza/traveler to download project
cd traveler/src
make build
cd ..
The binaries will be copied into traveler/bin. To navigate there from the src directory use: cd ../bin
-
Download the source code and
cdinto the traveler directory. -
Build an image:
docker build . -t traveler
- Run the container:
docker run -v "$PWD":/data -it traveler
The traveler executable is available in the PATH, and the current directory is mounted in the container under /data.
traveler [-h|--help]
traveler [OPTIONS] <STRUCTURES>
STRUCTURES:
<-gs|--target-structure> DBN_FILE
<-ts|--template-structure [--file-format FILE_FORMAT]> IMAGE_FILE DBN_FILE
DBN_FILE (Varna/DotBracketNotation) is in format like in example below
IMAGE_FILE* - visualization of template molecule, type of file can be specified by FILE_FORMAT argument
OPTIONS:
[-a|--all] [--overlaps] OUT_PREFIX
# computes mapping (TED) and outputs target leayout as both .ps and .svg image to files with prefix OUT_PREFIX
# with the optional --overlaps argument, overlaps in the layout are identified and highlited
[-t|--ted <FILE_MAPPING_OUT>]
# runs mapping (TED) only and saves mapping table to FILE_MAPPING_OUT file
[-d|--draw] [--overlaps] FILE_MAPPING_IN OUT_PREFIX
# use mapping in FILE_MAPPING_IN and outputs layout as both .ps and .svg image to files with prefix OUT_PREFIX
# if optional argument --overlaps is present overlaps in the layout are identified and highlighted
[-v|--verbose] Prints information about the computation and othere details (such as number of overlaps,
when overlap switch is turned on)
COLOR CODING:
Traveler uses the following color coding of nucleotides:
* Red - inserted bases
* Green - edited bases - e.g. the template has an adenosine at a position while the target has a cytosine at the same position and therefore cytosine will be colored green in the resulting layout)
* Blue - reinserted bases - happens when traveler needs to redraw simple structures like hairpins (for example due to the change in the number of bases)
* Brown - rotated parts - similar situation to reinserted bases, but takes place when redrawing a multibranch loop (in that case all branches need to be rotated to lie on a circle)
Three types of template IMAGE_FILE are currectly supported by Traveler:
- PostScript (crw) from CRW (default)
- VARNA (varna) format of SVG images produced by tool VARNA
- Traveler intermediate format
Traveler's intermediate format is a simple XML which contains an ordered list of nucleotides (information about lines is not necessary since lines connecting backbone are defined by the order of nucleotides and base pairing lines are defined by the secondary structure).
<structure>
<point x="138.3" y="-367" b="U" />
<point x="138.3" y="-359" b="A" />
.
.
.
<point x="278.1 y="-504.6" b="A" />
</structure>
Other extractors of RNA structure can be implemented and specified by the FILE_FORMAT argument.
Traveler accepts FASTA-like file format (see Example 0) for the description of the template structucture. You can prepare it manually, or, if you are using CRW as the source of templates, you can download the pseudoknot-free version version of the structure in the bpseq format and use it as the input to the bpseq2fasta Python script which can be found in the Utilities directory.
The file needs to contain three lines: moelcule description line (starts with the > symbol), sequence line, structure line. Should you have a sequence in a FASTA file with sequence spanning multiple lines, you can use the following script to obtain single-line sequence:
awk 'BEGIN {ORS=""}!/^>/{print}' sequence.fasta
$ cat data/metazoa/mouse.fasta
>mouse
UACCUGGUUGAUCCUGCCAGUAGCAUAUGCUUGUCUCAAAGAUUAAGCCAUGCAUGUCUAAGUACGCACGGCCGGUACAG
UGAAACUGCGAAUGGCUCAUUAAAUCAGUUAUGGUUCCUUUGGUCGCUCGCUCCUCUCCUACUUGGAUAACUGUGGUAAU
. . .
...(((((.......))))).((((((((((.(((((((((.....(((.(((..((...(((....((..........)
)...)))))......(((......((((..((..((....(((..................((((....(((((((....
. . .
FASTA (Varna/DBN) file should contain line starting with '>' and name of molecule, without any blank character
other lines are filled with LABELS and BRACKETS in dot-bracket notation of secondary structure pairing
match-tree must contain both LABELS and BRACKETS, templated-tree need only BRACKETS
Example 1a: Visualize mouse 18S rRNA using human 18S rRNA as template using CRW ps image as the template layout.
$ mkdir test
$ bin/traveler \
--target-structure data/metazoa/mouse.fasta \
--template-structure data/metazoa/human.ps data/metazoa/human.fasta \
--all test/mouse_from_human
Example 1b: Visualize mouse 18S rRNA using human 18S rRNA as template using Traveler's XML input format as the template layout.
$ mkdir test
$ bin/traveler \
--target-structure data/metazoa/mouse.fasta \
--template-structure --file-format traveler data/metazoa/human.xml data/metazoa/human.fasta \
--all test/mouse_from_human_xml_input
Example 2: Compute TED distance and mapping between human 18S rRNA (template) and mouse 18S rRNA (target).
$ mkdir test
$ bin/traveler \
--target-structure data/metazoa/mouse.fasta \
--template-structure data/metazoa/human.ps data/metazoa/human.fasta \
--ted test/mouse_from_human.map
$ mkdir test
$ bin/traveler \
--target-structure data/metazoa/mouse.fasta \
--template-structure data/metazoa/human.ps data/metazoa/human.fasta \
--draw --overlaps test/mouse_from_human.map test/mouse_from_human
$ # generates 4 files - .svg and .ps files, both with/without colored bases (see COLOR CODING section)
$ # checks also if output molecule has overlaps and draws them in output image
Options --ted and --draw serve for separatation of mapping and visualization since TED computation and on the other hand, Traveler allows for multiple output visualization (coloring, overlaps).
Traveler supporst two types of input images - crw and varna. There are three steps that need to done when one wants to support other image types.
- You need to implement
extractorinterface and its methodextract. The method accepts nucleotides (the primary structure) and their position in the image (points) from given file. - In the class, you need to implement method
get_type, that should only return extractor's type that is used in IMAGE_FILE argument. - Add your new extractor to method
extractor.get_all_extractors()
For more ideas, how it should be implemented, see usage of crw_extractor and varna_extractor.
If you use Traveler in your research, please cite:
- Elias, R., & Hoksza, D. (2017). TRAVeLer: a tool for template-based RNA secondary structure visualization. BMC bioinformatics, 18(1), 487.
- Eliás, R., & Hoksza, D. (2016). Rna secondary structure visualization using tree edit distance. International Journal of Bioscience, Biochemistry and Bioinformatics, 6(1), 9.