Dynamax is a library for probabilistic state space models (SSMs) written in JAX. It has code for inference (state estimation) and learning (parameter estimation) in a variety of SSMs, including:
- Hidden Markov Models (HMMs)
- Linear Gaussian State Space Models (aka Linear Dynamical Systems)
- Nonlinear Gaussian State Space Models
- Generalized Gaussian State Space Models (with non-Gaussian emission models)
The library consists of a set of core, functionally pure, low-level inference algorithms, as well as a set of model classes which provide a more user-friendly, object-oriented interface. It is compatible with other libraries in the JAX ecosystem, such as optax (used for estimating parameters using stochastic gradient descent), and Blackjax (used for computing the parameter posterior using Hamiltonian Monte Carlo (HMC) or sequential Monte Carlo (SMC)).
For tutorials and API documentation, see: https://probml.github.io/dynamax/.
To install the latest releast of dynamax from PyPi:
pip install dynamax # Install dynamax and core dependencies, or
pip install dynamax[notebooks] # Install with demo notebook dependencies
To install the latest development branch:
pip install git+https://github.com/probml/dynamax.git
Finally, if you're a developer, you can install dynamax along with the test and documentation dependencies with:
git clone [email protected]:probml/dynamax.git
cd dynamax
pip install -e '.[dev]'
To run the tests:
pytest dynamax # Run all tests
pytest dynamax/hmm/inference_test.py # Run a specific test
pytest -k lgssm # Run tests with lgssm in the name
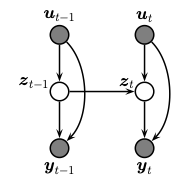
A state space model or SSM is a partially observed Markov model, in
which the hidden state,
The corresponding joint distribution has the following form (in dynamax, we restrict attention to discrete time systems):
Here
We assume that we see the observations
More information can be found in these books:
- "Machine Learning: Advanced Topics", K. Murphy, MIT Press 2023. Available at https://probml.github.io/pml-book/book2.html.
- "Bayesian Filtering and Smoothing", S. Särkkä, Cambridge University Press, 2013. Available at https://users.aalto.fi/~ssarkka/pub/cup_book_online_20131111.pdf
Dynamax includes classes for many kinds of SSM. You can use these models to simulate data, and you can fit the models using standard learning algorithms like expectation-maximization (EM) and stochastic gradient descent (SGD). Below we illustrate the high level (object-oriented) API for the case of an HMM with Gaussian emissions. (See this notebook for a runnable version of this code.)
import jax.numpy as jnp
import jax.random as jr
import matplotlib.pyplot as plt
from dynamax.hidden_markov_model import GaussianHMM
key1, key2, key3 = jr.split(jr.PRNGKey(0), 3)
num_states = 3
emission_dim = 2
num_timesteps = 1000
# Make a Gaussian HMM and sample data from it
hmm = GaussianHMM(num_states, emission_dim)
true_params, _ = hmm.initialize(key1)
true_states, emissions = hmm.sample(true_params, key2, num_timesteps)
# Make a new Gaussian HMM and fit it with EM
params, props = hmm.initialize(key3, method="kmeans", emissions=emissions)
params, lls = hmm.fit_em(params, props, emissions, num_iters=20)
# Plot the marginal log probs across EM iterations
plt.plot(lls)
plt.xlabel("EM iterations")
plt.ylabel("marginal log prob.")
# Use fitted model for posterior inference
post = hmm.smoother(params, emissions)
print(post.smoothed_probs.shape) # (1000, 3)JAX allows you to easily vectorize these operations with vmap.
For example, you can sample and fit to a batch of emissions as shown below.
from functools import partial
from jax import vmap
num_seq = 200
batch_true_states, batch_emissions = \
vmap(partial(hmm.sample, true_params, num_timesteps=num_timesteps))(
jr.split(key2, num_seq))
print(batch_true_states.shape, batch_emissions.shape) # (200,1000) and (200,1000,2)
# Make a new Gaussian HMM and fit it with EM
params, props = hmm.initialize(key3, method="kmeans", emissions=batch_emissions)
params, lls = hmm.fit_em(params, props, batch_emissions, num_iters=20)These examples demonstrate the dynamax models, but we can also call the low-level inference code directly.
Please see this page for details on how to contribute.
Core team: Peter Chang, Giles Harper-Donnelly, Aleyna Kara, Xinglong Li, Scott Linderman, Kevin Murphy.
Other contributors: Adrien Corenflos, Elizabeth DuPre, Gerardo Duran-Martin, Colin Schlager, Libby Zhang and other people listed here
MIT License. 2022