-
-
Notifications
You must be signed in to change notification settings - Fork 1.1k
New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
Chunked processing across multiple raster (geoTIF) files #2314
Comments
|
Can you provide a |
|
I think the explicit
|
|
As @darothen mentioned, first thing is to check that the geotiffs themselves are tiled (otherwise I'm guessing that open_rasterio() will open the entire thing. You can do this with: import rasterio
with rasterio.open('image_001.tif') as src:
print(src.profile)Here is the mentioned example notebook which works for tiled geotiffs stored on google cloud: You can use the 'launch binder' button to run it with a pangeo dask-kubernetes cluster, or just read through the landsat8-cog-ndvi.ipynb notebook. |
|
I see now that you are using dask-distributed, but I guess there are still too many intermediate outputs here to do a single rechunk operation. The crude but effective way to solve this problem would be to loop over spatial tiles using an indexing operation to pull out only a limited extent, compute the calculation on each tile and then reassemble the tiles at the end. To see if this will work, you might try computing a single time-series on your merged dataset before calling In theory, I think using |
|
Thanks for all the suggestions! An update from when I originally posted this. Aligning with @shoyer, @darothen and @scottyhq 's comments, we've tested the code using cloud-optimized geoTIF files and regular geoTIFs, and it does perform better with the cloud-optimized form, though still appears to "over-eagerly" load more than just what is being worked on. With the cloud-optimized form, performance is much better when we specify the chunking strategy on the initial open_rasterio and it aligns with the chunk sizes. The result is a larger task graph (and much more time spent developing the task graph) but more cases where we don't run into memory problems. There still appears to be a lot more memory used than I expect, but am actively working on exploring options. We've also noticed better performance using a k8s Dask cluster distributed across multiple "independent" workers as opposed to using a LocalCluster on a single large machine. As in, with the distributed cluster the "myfunction" (fit) operation starts happening on chunks well before the entire dataset is loaded, whereas in the LocalCluster it still tends not to begin until all chunks have been loaded in. Not exactly sure what would cause that... I'm intrigued by @shoyer 's last suggestion of an "intermediate" chunking step. Will test that and potentially the manual iteration over the tiles. Thanks for all the suggestions and thoughts! |
|
Has there been any progress on this issue? I am bumping into the same problem. |
|
This particular use case is extremely common when working with spatio-temporal data. Can anyone suggest a good workaround for this? |
|
Hi @shaprann, I haven't re-visited this exact workflow recently, but one really good option (if you can manage the intermediate storage cost) would be to try to use new tools like http://github.com/pangeo-data/rechunker to pre-process and prepare your data archive prior to analysis. |
|
Just noticed this issue; people needing to do this sort of thing might want to look at stackstac (especially playing with the
FYI, this is basically expected behavior for distributed, see: |
|
We've deleted the internal |
Looking for guidance on best practices, or forcing dask/xarray to work in the way I want it to work :)
Task: I have hundreds of geoTIF files (500 for this test case), each containing a single 2d x-y array (~10000 x 10000). What I need to do is read these all in and apply a function to each x-y point whose argument is a 1-d array populated by the data from that same x-y point across all the files. This function is an optimization fit at each x-y point, so it's not something easily vectorizable, but each point's calculation is independent.
Because each point may be treated independently, the idea is to be able to split the dataset into chunks in the x-y directions and allow Dask to cycle through and fork out the tasks of applying the function to each chunk in turn. Here's how I'm currently trying to implement this:
Problem description
What happens now is that all the raster datasets appear to be read into memory in full immediately and without respect to the final chunk alignment needed for the calculation. Then, there seems to be a lot of memory usage and communications overhead as the different chunks get rectified in the concatenation and merge-rechunk operations before myfunction is applied. This blows up the memory requirements, exceeding 300 GB RAM on a 48 worker machine (dataset on disk is ~100 GB). Below is a screenshot of the Dask dashboard right before workers start cutting out with memory limits/can't dump to disk errors.
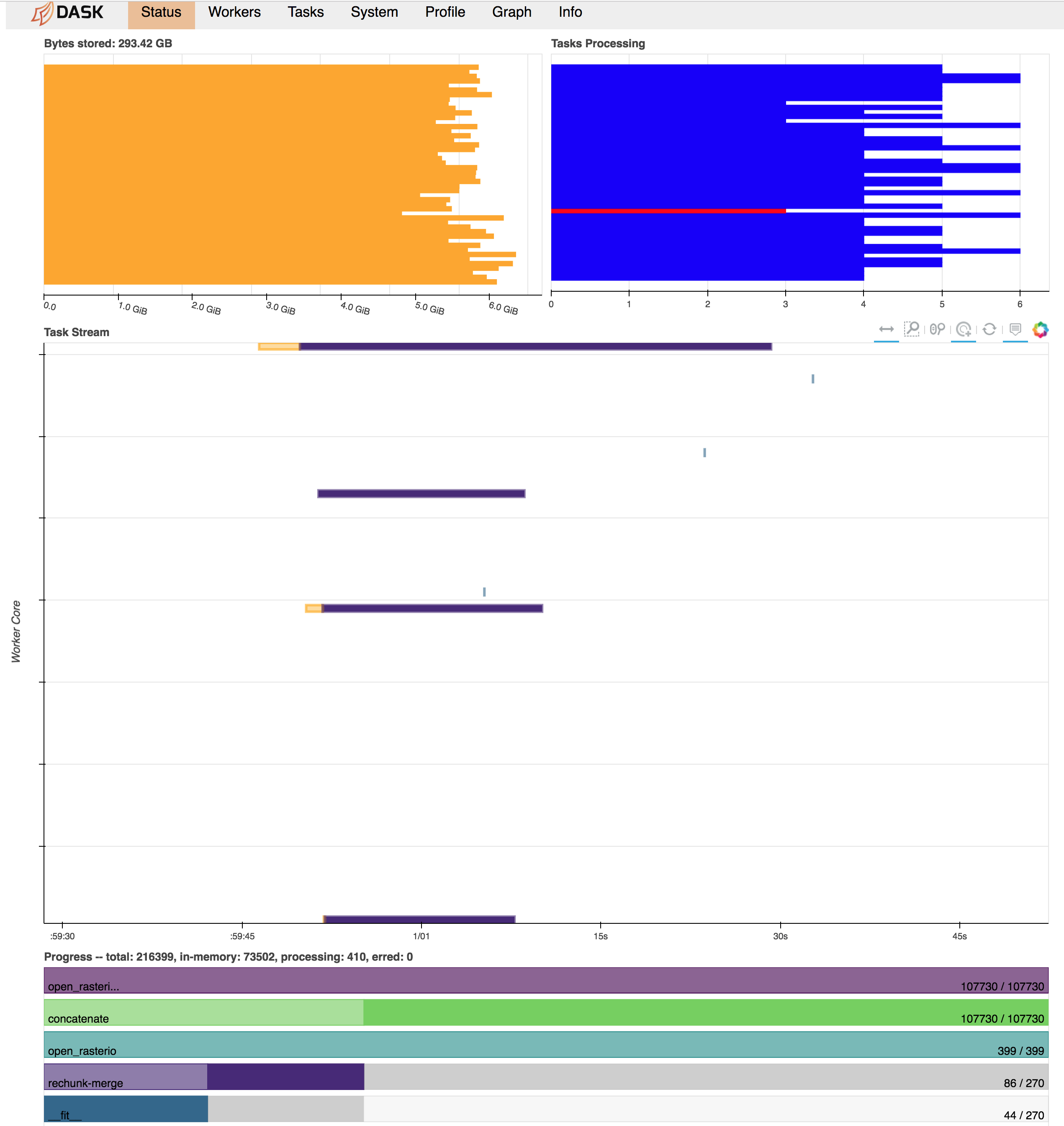
Expected Output
What I'm looking for in this is an out-of-core computation where a single worker stays with a single chunk of the data all the way through to the end. Each "task" would then be a sequence of:
I could manually sort this all out with a bunch of dask.delayed tasks, keeping track of the indices/locations of each chunk as they were returned to "reassemble" them later. However, is there an easier way to simplify this operation directly through xarray/dask through some ordering of commands I haven't found yet? Does the netcdf open_mfdataset function somehow handle this sort of idea for multiple netcdf files that could be adapted for rasterio?
Output of
xr.show_versions()xarray: 0.10.7
pandas: 0.23.2
numpy: 1.14.5
scipy: 1.1.0
netCDF4: 1.4.0
h5netcdf: 0.6.1
h5py: 2.8.0
Nio: None
zarr: 2.2.0
bottleneck: 1.2.1
cyordereddict: None
dask: 0.18.1
distributed: 1.22.0
matplotlib: 2.2.2
cartopy: None
seaborn: None
setuptools: 36.5.0.post20170921
pip: 9.0.1
conda: 4.5.5
pytest: None
IPython: 6.4.0
sphinx: None
The text was updated successfully, but these errors were encountered: