chemcloud is a python client for the ChemCloud Server. The client provides a simple yet powerful interface to perform computational chemistry calculations at scale using nothing but modern Python and an internet connection.
Documentation: https://mtzgroup.github.io/chemcloud-client
chemcloud works in harmony with a suite of other quantum chemistry tools for fast, structured, and interoperable quantum chemistry.
- qcio - Elegant and intuitive data structures for quantum chemistry, featuring seamless Jupyter Notebook visualizations. Documentation
- qcparse - A library for efficient parsing of quantum chemistry data into structured
qcioobjects. - qcop - A package for operating quantum chemistry programs using
qciostandardized data structures. Compatible withTeraChem,psi4,QChem,NWChem,ORCA,Molpro,geomeTRIC, and many more, featuring seamless Jupyter Notebook visualizations. - BigChem - A distributed application for running quantum chemistry calculations at scale across clusters of computers or the cloud. Bring multi-node scaling to your favorite quantum chemistry program, featuring seamless Jupyter Notebook visualizations.
ChemCloud- A web application and associated Python client for exposing a BigChem cluster securely over the internet, featuring seamless Jupyter Notebook visualizations.
pip install chemcloudRun calculations just like you would with qcop except calling chemcloud.compute instead of qcop.compute. You may also pass list of inputs to chemcloud.compute to run calculations in parallel. By default chemcloud.compute will return ProgramOutput objects for all calculations, even those that failed, rather than raising exceptions. Check if calculations were successful by accessing output.success.
from qcio import Structure, ProgramInput
from chemcloud import compute
# Create the structure
h2o = Structure.open("h2o.xyz")
# Define the program input
prog_input = ProgramInput(
structure=h2o,
calctype="energy",
model={"method": "hf", "basis": "sto-3g"},
keywords={"purify": "no", "restricted": False},
)
# Submit the calculation to the server
output = compute("terachem", prog_input)
# Inspect the output
output.input_data # Input data used by the QC program
output.success # Whether the calculation succeeded
output.results # All structured results from the calculation
output.stdout # Stdout log from the calculation
output.pstdout # Shortcut to print out the stdout in human readable format
output.files # Any files returned by the calculation
output.provenance # Provenance information about the calculation
output.extras # Any extra information not in the schema
output.traceback # Stack trace if calculation failed
output.ptraceback # Shortcut to print out the traceback in human readable formatSubmit thousands of calculations simultaneously and collect results parallel:
prog_inputs = [prog_input] * 10
outputs = compute("terachem", prog_inputs)
for output in outputs:
# Process outputs
output.save(...)Or stream results from the server as they complete:
prog_inputs = [prog_input] * 10
# Submit the calculation to the server
future = compute("terachem", prog_inputs, return_future=True)
for output in future.as_completed():
# Outputs returned as they complete
output.save(...)If you want to use a non-blocking API, pass return_future=True to compute. Calling .get() on the future will return a ProgramOutput or list of ProgramOutput once the calculations are complete.
prog_inputs = [prog_input] * 10
# Submit the calculation to the server
future = compute("terachem", prog_inputs, return_future=True)
# Check the status of calculations (optional)
future.is_ready
# Block and retrieve results
outputs = future.get()
for output in outputs:
# Process outputs
output.save(...)Save a future to disk and then collect results later:
# Submit the calculation to the server
future = compute("terachem", prog_inputs, return_future=True)
future.save("myfuture.json")
# Later in a different script
future.open("myfuture.json")
outputs = future.get()More examples can be found in the examples directory.
Visualize all your results with a single line of code!
First install the visualization module:
pip install qcio[view]or if your shell requires '' around arguments with brackets:
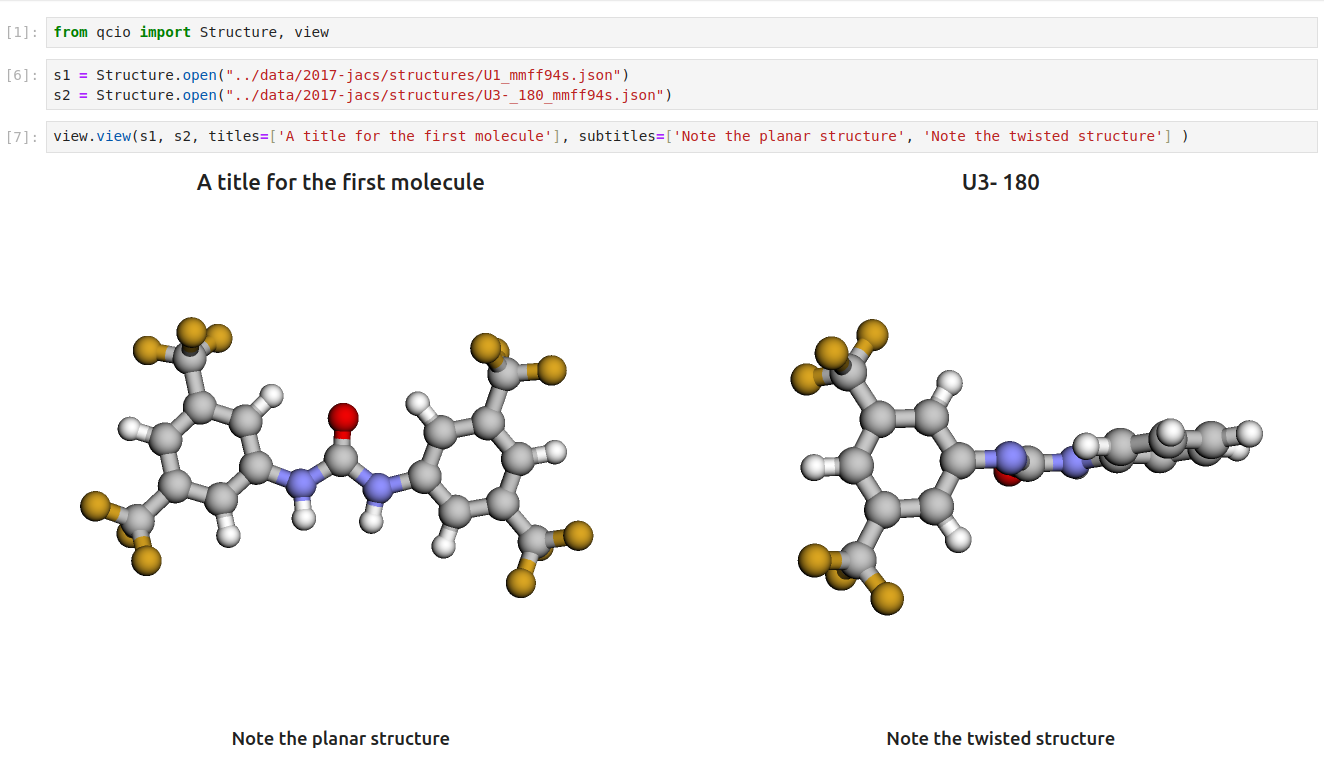
pip install 'qcio[view]'Then in a Jupyter notebook import the qcio view module and call view.view(...) passing it one or any number of qcio objects you want to visualizing including Structure objects or any ProgramOutput object. You may also pass an array of titles and/or subtitles to add additional information to the molecular structure display. If no titles are passed qcio with look for Structure identifiers such as a name or SMILES to label the Structure.
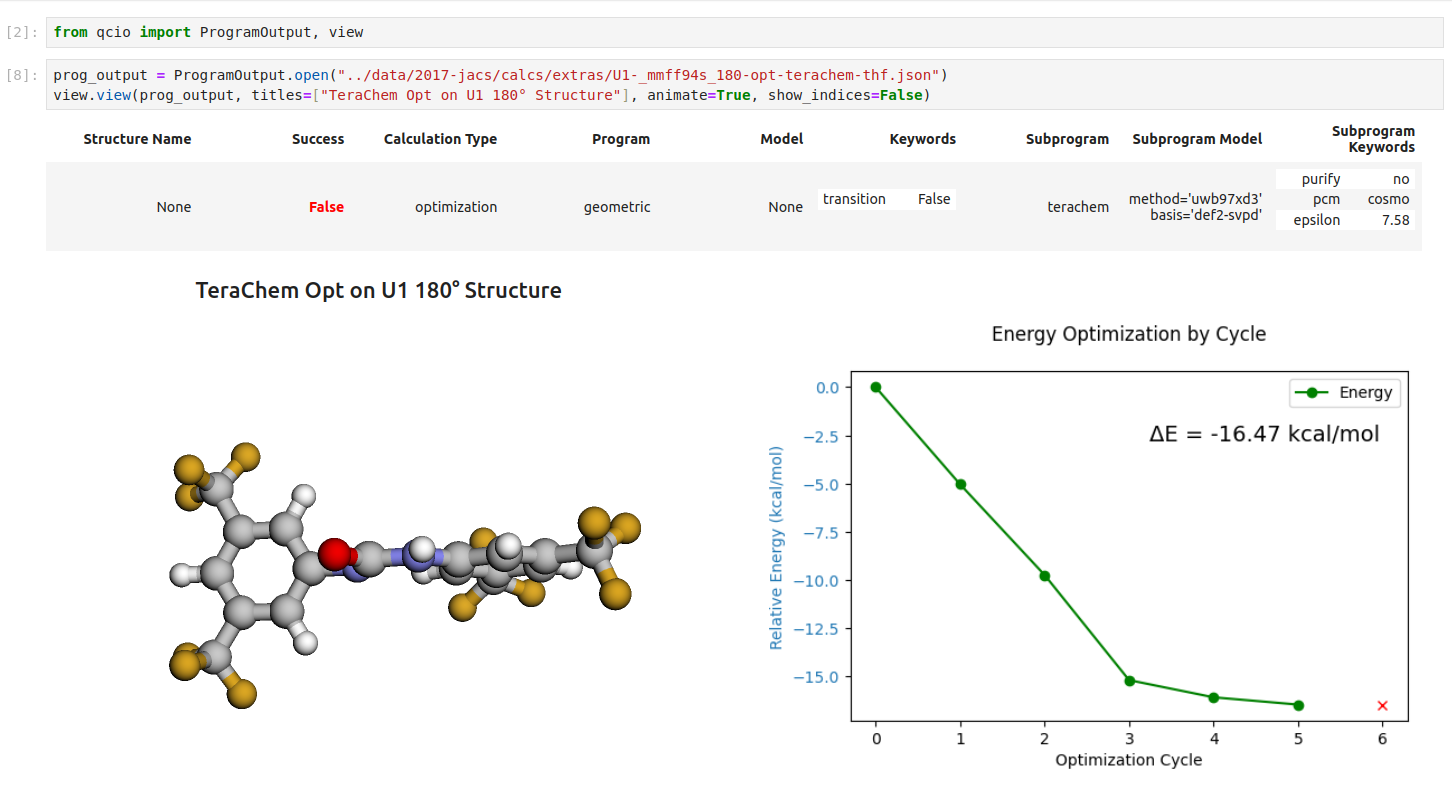
Seamless visualizations for ProgramOutput objects make results analysis easy!
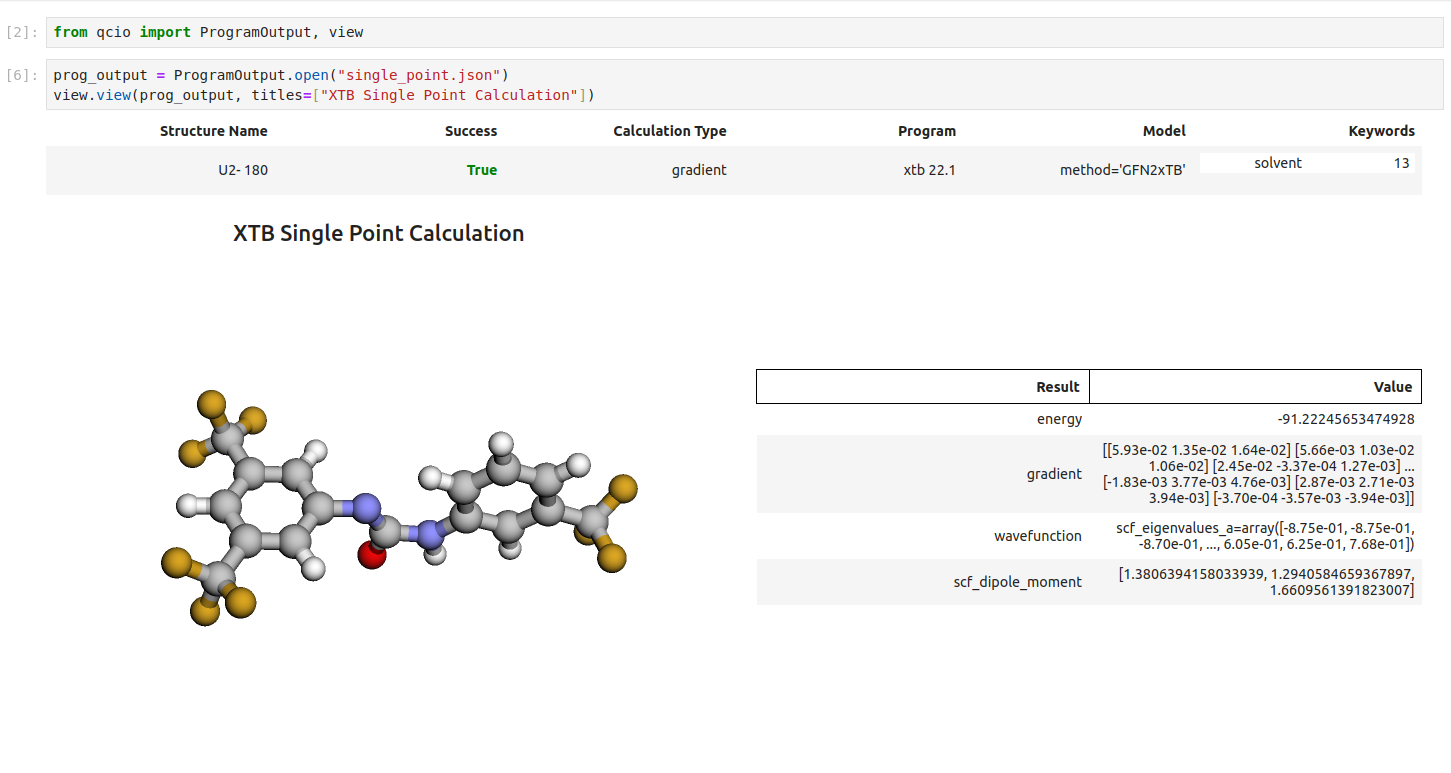
Single point calculations display their results in a table.
If you want to use the HTML generated by the viewer to build your own dashboards use the functions inside of qcio.view.py that begin with the word generate_ to create HTML you can insert into any dashboard.
If you have any issues with chemcloud or would like to request a feature, please open an issue.