MATLAB code for dimensionality reduction, fault detection, and fault diagnosis using KPCA
Version 2.2, 14-MAY-2021
Email: [email protected]
- Easy-used API for training and testing KPCA model
- Support for dimensionality reduction, data reconstruction, fault detection, and fault diagnosis
- Multiple kinds of kernel functions (linear, gaussian, polynomial, sigmoid, laplacian)
- Visualization of training and test results
- Component number determination based on given explained level or given number
- Only fault diagnosis of Gaussian kernel is supported.
- This code is for reference only.
A class named Kernel is defined to compute kernel function matrix.
%{
type -
linear : k(x,y) = x'*y
polynomial : k(x,y) = (γ*x'*y+c)^d
gaussian : k(x,y) = exp(-γ*||x-y||^2)
sigmoid : k(x,y) = tanh(γ*x'*y+c)
laplacian : k(x,y) = exp(-γ*||x-y||)
degree - d
offset - c
gamma - γ
%}
kernel = Kernel('type', 'gaussian', 'gamma', value);
kernel = Kernel('type', 'polynomial', 'degree', value);
kernel = Kernel('type', 'linear');
kernel = Kernel('type', 'sigmoid', 'gamma', value);
kernel = Kernel('type', 'laplacian', 'gamma', value);For example, compute the kernel matrix between X and Y
X = rand(5, 2);
Y = rand(3, 2);
kernel = Kernel('type', 'gaussian', 'gamma', 2);
kernelMatrix = kernel.computeMatrix(X, Y);
>> kernelMatrix
kernelMatrix =
0.5684 0.5607 0.4007
0.4651 0.8383 0.5091
0.8392 0.7116 0.9834
0.4731 0.8816 0.8052
0.5034 0.9807 0.7274clc
clear all
close all
addpath(genpath(pwd))
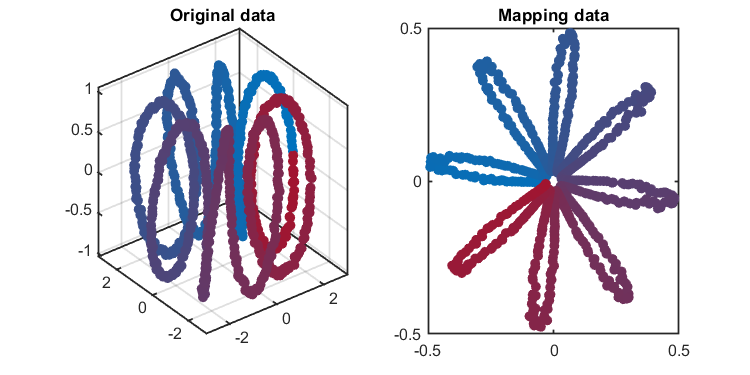
load('.\data\helix.mat', 'data')
kernel = Kernel('type', 'gaussian', 'gamma', 2);
parameter = struct('numComponents', 2, ...
'kernelFunc', kernel);
% build a KPCA object
kpca = KernelPCA(parameter);
% train KPCA model
kpca.train(data);
% mapping data
mappingData = kpca.score;
% Visualization
kplot = KernelPCAVisualization();
% visulize the mapping data
kplot.score(kpca)The training results (dimensionality reduction):
*** KPCA model training finished ***
running time = 0.2798 seconds
kernel function = gaussian
number of samples = 1000
number of features = 3
number of components = 2
number of T2 alarm = 135
number of SPE alarm = 0
accuracy of T2 = 86.5000%
accuracy of SPE = 100.0000% Another application using banana-shaped data:
clc
clear all
close all
addpath(genpath(pwd))
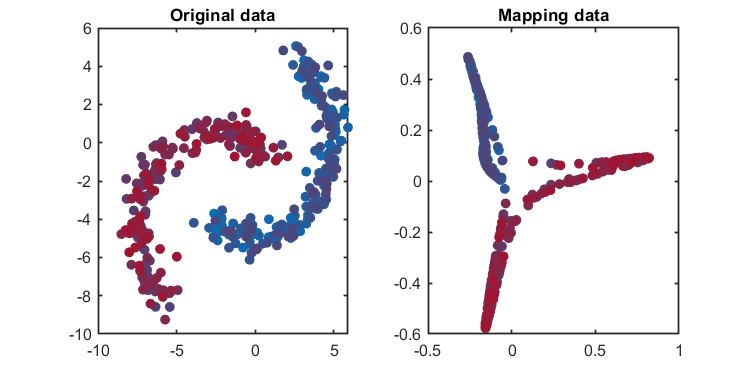
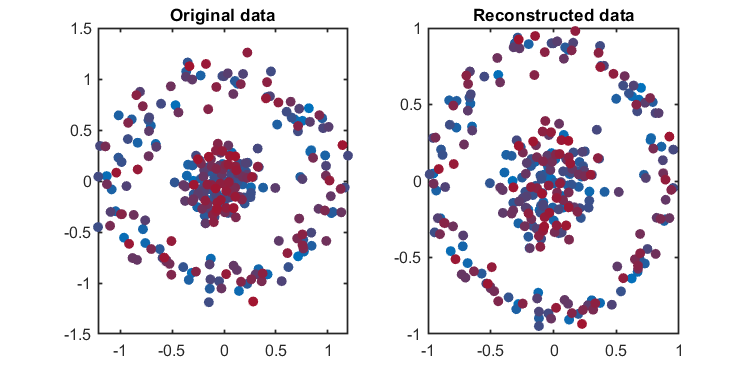
load('.\data\circle.mat', 'data')
kernel = Kernel('type', 'gaussian', 'gamma', 0.2);
parameter = struct('numComponents', 2, ...
'kernelFunc', kernel);
% build a KPCA object
kpca = KernelPCA(parameter);
% train KPCA model
kpca.train(data);
% reconstructed data
reconstructedData = kpca.newData;
% Visualization
kplot = KernelPCAVisualization();
kplot.reconstruction(kpca)The Component number can be determined based on given explained level or given number.
Case 1
The number of components is determined by the given explained level. The given explained level should be 0 < explained level < 1. For example, when explained level is set to 0.75, the parameter should be set as:
parameter = struct('numComponents', 0.75, ...
'kernelFunc', kernel);The code is
clc
clear all
close all
addpath(genpath(pwd))
load('.\data\TE.mat', 'trainData')
kernel = Kernel('type', 'gaussian', 'gamma', 1/128^2);
parameter = struct('numComponents', 0.75, ...
'kernelFunc', kernel);
% build a KPCA object
kpca = KernelPCA(parameter);
% train KPCA model
kpca.train(trainData);
% Visualization
kplot = KernelPCAVisualization();
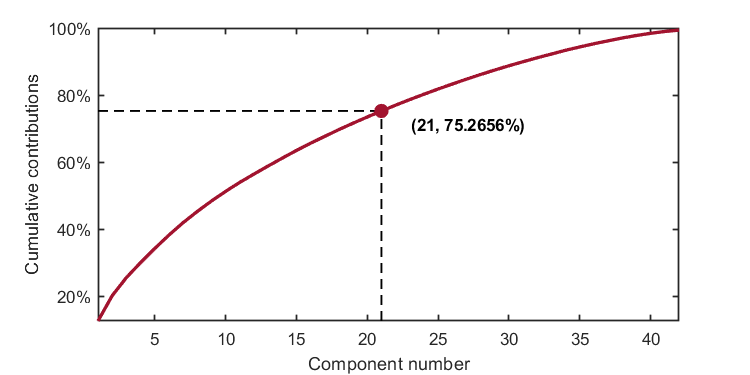
kplot.cumContribution(kpca)As shown in the image, when the number of components is 21, the cumulative contribution rate is 75.2656%,which exceeds the given explained level (0.75).
Case 2
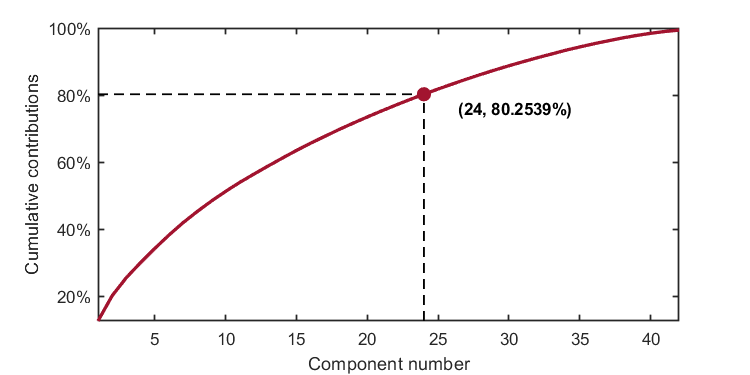
The number of components is determined by the given number. For example, when the given number is set to 24, the parameter should be set as:
parameter = struct('numComponents', 24, ...
'kernelFunc', kernel);The code is
clc
clear all
close all
addpath(genpath(pwd))
load('.\data\TE.mat', 'trainData')
kernel = Kernel('type', 'gaussian', 'gamma', 1/128^2);
parameter = struct('numComponents', 24, ...
'kernelFunc', kernel);
% build a KPCA object
kpca = KernelPCA(parameter);
% train KPCA model
kpca.train(trainData);
% Visualization
kplot = KernelPCAVisualization();
kplot.cumContribution(kpca)As shown in the image, when the number of components is 24, the cumulative contribution rate is 80.2539%.
Demonstration of fault detection using KPCA (TE process data)
clc
clear all
close all
addpath(genpath(pwd))
load('.\data\TE.mat', 'trainData', 'testData')
kernel = Kernel('type', 'gaussian', 'gamma', 1/128^2);
parameter = struct('numComponents', 0.65, ...
'kernelFunc', kernel);
% build a KPCA object
kpca = KernelPCA(parameter);
% train KPCA model
kpca.train(trainData);
% test KPCA model
results = kpca.test(testData);
% Visualization
kplot = KernelPCAVisualization();
kplot.cumContribution(kpca)
kplot.trainResults(kpca)
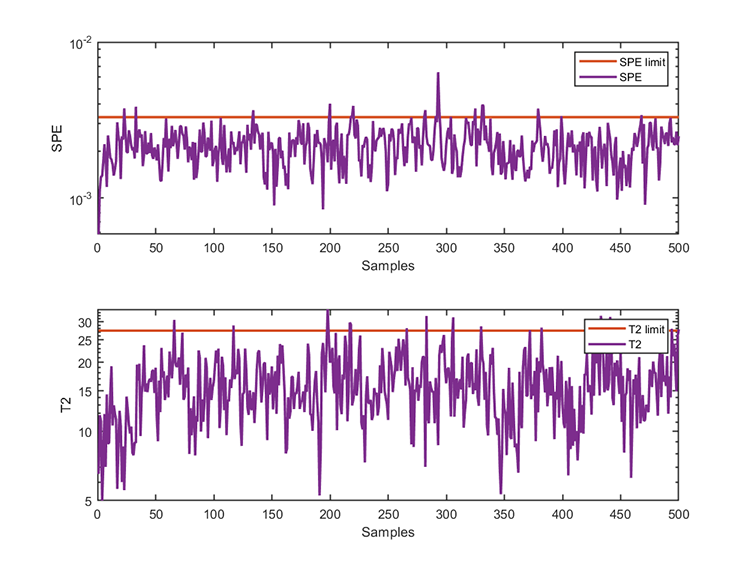
kplot.testResults(kpca, results)The training results are
*** KPCA model training finished ***
running time = 0.0986 seconds
kernel function = gaussian
number of samples = 500
number of features = 52
number of components = 16
number of T2 alarm = 16
number of SPE alarm = 17
accuracy of T2 = 96.8000%
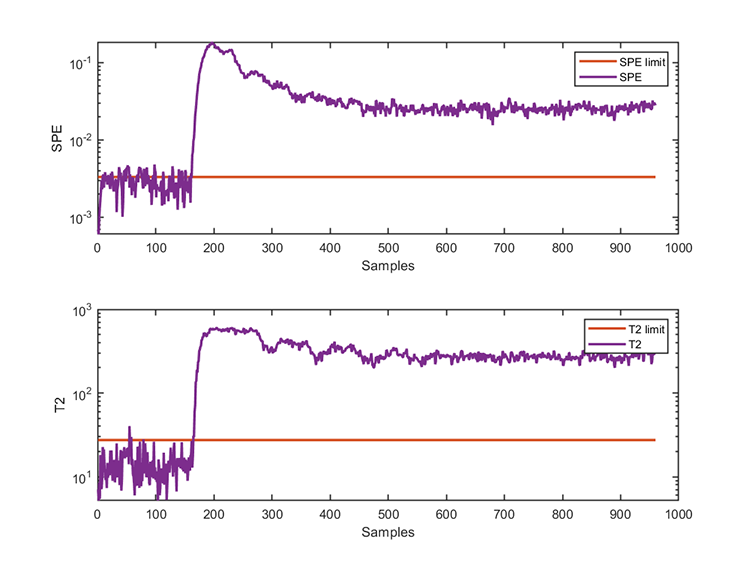
accuracy of SPE = 96.6000% The test results are
*** KPCA model test finished ***
running time = 0.0312 seconds
number of test data = 960
number of T2 alarm = 799
number of SPE alarm = 851 Notice
- If you want to calculate CPS of a certain time, you should set starting time equal to ending time. For example, 'diagnosis', [500, 500]
- If you want to calculate the average CPS of a period of time, starting time and ending time should be set respectively. 'diagnosis', [300, 500]
- The fault diagnosis module is only supported for gaussian kernel function and it may still take a long time when the number of the training data is large.
clc
clear all
close all
addpath(genpath(pwd))
load('.\data\TE.mat', 'trainData', 'testData')
kernel = Kernel('type', 'gaussian', 'gamma', 1/128^2);
parameter = struct('numComponents', 0.65, ...
'kernelFunc', kernel,...
'diagnosis', [300, 500]);
% build a KPCA object
kpca = KernelPCA(parameter);
% train KPCA model
kpca.train(trainData);
% test KPCA model
results = kpca.test(testData);
% Visualization
kplot = KernelPCAVisualization();
kplot.cumContribution(kpca)
kplot.trainResults(kpca)
kplot.testResults(kpca, results)
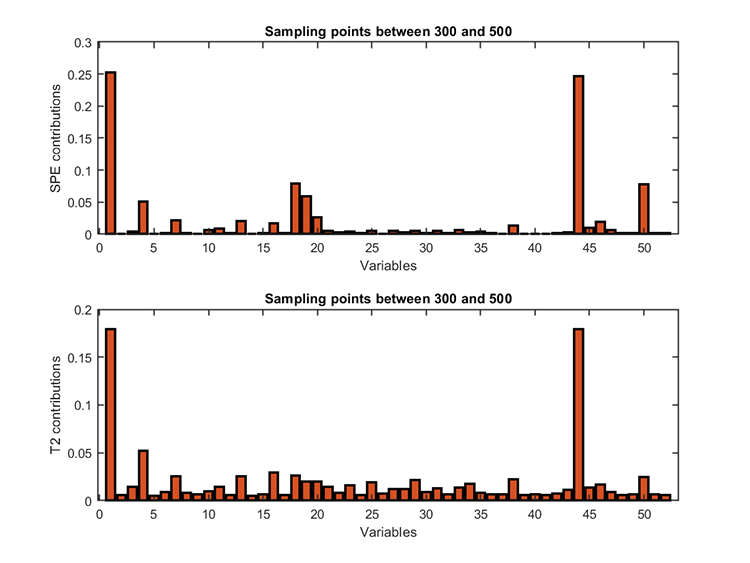
kplot.diagnosis(results)Diagnosis results:
*** Fault diagnosis ***
Fault diagnosis start...
Fault diagnosis finished.
running time = 18.2738 seconds
start point = 300
ending point = 500
fault variables (T2) = 44 1 4
fault variables (SPE) = 1 44 18