diff --git a/CONTRIBUTORS.yaml b/CONTRIBUTORS.yaml
index 056c097c4d8fbb..8a9a770e48130e 100644
--- a/CONTRIBUTORS.yaml
+++ b/CONTRIBUTORS.yaml
@@ -51,6 +51,7 @@ awspolly:
# our real contributors <3 (please add yourself in alphabetical order)
+
a-asai:
name: Atsushi Asai
email: tbudcm7zk@i.softbank.jp
@@ -86,6 +87,14 @@ abueg:
orcid: 0000-0002-6879-3954
joined: 2022-01
+adya29:
+ name: Adya Shreya
+ email: adya_shreya@gis.a-star.edu.sg
+ joined: 2024-12
+ linkedin: adya-shreya
+ location:
+ country: SG
+
adairama:
name: Adaikalavan Ramasamy
email: adai@gis.a-star.edu.sg
@@ -95,6 +104,7 @@ adairama:
location:
country: SG
+
ahmedhamidawan:
name: Ahmed Hamid Awan
email: aawan7@jhu.edu
diff --git a/events/2022-07-08-gat.md b/events/2022-07-08-gat.md
index 6f4dd17d32a19b..b986234f4c5cca 100644
--- a/events/2022-07-08-gat.md
+++ b/events/2022-07-08-gat.md
@@ -119,7 +119,7 @@ program:
topic: admin
- name: tool-integration
topic: dev
- - name: processing-many-samples-at-once
+ - name: collections
topic: galaxy-interface
- name: upload-rules
topic: galaxy-interface
diff --git a/events/2023-04-17-gat-gent.md b/events/2023-04-17-gat-gent.md
index 145886c08bb5a5..bea47e80fb06e3 100644
--- a/events/2023-04-17-gat-gent.md
+++ b/events/2023-04-17-gat-gent.md
@@ -223,7 +223,7 @@ program:
time: "11:30 - 11:45"
- name: tool-integration
topic: dev
- - name: processing-many-samples-at-once
+ - name: collections
topic: galaxy-interface
- name: upload-rules
topic: galaxy-interface
diff --git a/faqs/galaxy/visualisations_igv.md b/faqs/galaxy/visualisations_igv.md
index 7f7119795b9a83..b0b654ed0f7231 100644
--- a/faqs/galaxy/visualisations_igv.md
+++ b/faqs/galaxy/visualisations_igv.md
@@ -4,6 +4,7 @@ area: visualisation
box_type: tip
layout: faq
contributors: [shiltemann]
+redirect_from: [/topics/galaxy-interface/tutorials/processing-many-samples-at-once/faqs/visualisations_igv]
---
You can send data from your Galaxy history to IGV for viewing as follows:
diff --git a/learning-pathways/admin-training.md b/learning-pathways/admin-training.md
index 46d484b6124196..436cc5c9110f3e 100644
--- a/learning-pathways/admin-training.md
+++ b/learning-pathways/admin-training.md
@@ -88,7 +88,7 @@ pathway:
topic: admin
- name: tool-integration
topic: dev
- - name: processing-many-samples-at-once
+ - name: collections
topic: galaxy-interface
- name: upload-rules
topic: galaxy-interface
diff --git a/news/_posts/2024-12-06-spoc_cofest.md b/news/_posts/2024-12-06-spoc_cofest.md
new file mode 100644
index 00000000000000..cd9f3be5a6194a
--- /dev/null
+++ b/news/_posts/2024-12-06-spoc_cofest.md
@@ -0,0 +1,19 @@
+---
+title: "SPOC CoFest 2024: How did it go?"
+contributions:
+ authorship: [nomadscientist]
+tags: [gtn, single-cell]
+layout: news
+cover: "news/images/2023_dec_sc.png"
+coveralt: "swirled cluster dots surround a circle of people all holding hands, looking towards the bright center (future)"
+---
+
+# First SPOC CoFest
+
+We held our first 🖖🏾[SPOC CoFest]({% link events/2024-12-06-spoc-cofest-2024.md %}), in the great tradition of the excellent CoFests organised in the GTN that welcomed @gtn:nomadscientist and many others into the community.
+
+We welcomed a diverse group of participants, from experienced Galaxy trainers to bioinformaticians with little to no Galaxy experience whatsoever. We also were lucky enough to be joined by a 5-strong outpost from Singapore, prompting a Pacific-time-zone kick-off session prior to our day-long event.
+
+🎉A big thank you to all the participants for their hard work!🎉
+
+
diff --git a/news/images/2024-12-06-spoc-cofest_outputs.png b/news/images/2024-12-06-spoc-cofest_outputs.png
new file mode 100644
index 00000000000000..f672f56b009806
Binary files /dev/null and b/news/images/2024-12-06-spoc-cofest_outputs.png differ
diff --git a/topics/admin/tutorials/monitoring/tutorial.md b/topics/admin/tutorials/monitoring/tutorial.md
index 1a8fdc02e49a85..785260c51044bc 100644
--- a/topics/admin/tutorials/monitoring/tutorial.md
+++ b/topics/admin/tutorials/monitoring/tutorial.md
@@ -364,7 +364,7 @@ Setting up Telegraf is again very simple. We just add a single role to our playb
> + - plugin: disk
> + - plugin: kernel
> + - plugin: processes
-> + - plugin: io
+> + - plugin: diskio
> + - plugin: mem
> + - plugin: system
> + - plugin: swap
diff --git a/topics/ecology/tutorials/Metashrimps_tutorial/tutorial.md b/topics/ecology/tutorials/Metashrimps_tutorial/tutorial.md
index ae49d6024dc5b1..b7bf1e2f47bd5d 100644
--- a/topics/ecology/tutorials/Metashrimps_tutorial/tutorial.md
+++ b/topics/ecology/tutorials/Metashrimps_tutorial/tutorial.md
@@ -86,7 +86,7 @@ publishable as a real Data Paper giving recognition to all the people that helpe
>
> 1. Create a new history for this tutorial
> 2. Import this metadata file from [Zenodo]({{ page.zenodo_link }}) to test it
-> -> Training Data for "Creating Quality FAIR assessment reports and draft of Data Papers from EML metadata with MetaShRIMPS"):
+> -> Training Data for "Creating Quality FAIR assessment reports and draft of Data Papers from EML metadata with MetaShRIMPS":
> ```
> https://zenodo.org/record/8130567/files/Kakila_database_marine_mammal.xml
> ```
@@ -109,7 +109,7 @@ you want to use.
{:width="500"}
-After uploading the file, or if you have indicate it as input data if the tool, you just have to click on **Execute** to launch the tool with the file.
+After uploading the file, or if you have indicated it as input data in the tool form, you just have to click on **Execute** to launch MetaShRIMPS jobs with the file.
# Outputs
diff --git a/topics/galaxy-interface/metadata.yaml b/topics/galaxy-interface/metadata.yaml
index 5071040f57f695..b8a0078ee11261 100644
--- a/topics/galaxy-interface/metadata.yaml
+++ b/topics/galaxy-interface/metadata.yaml
@@ -5,6 +5,11 @@ title: "Using Galaxy and Managing your Data"
summary: "A collection of microtutorials explaining various features of the Galaxy user interface and manipulating data within Galaxy."
docker_image:
requirements:
+ -
+ type: "internal"
+ topic_name: introduction
+ tutorials:
+ - galaxy-intro-101
subtopics:
- id: upload
diff --git a/topics/galaxy-interface/tutorials/collections/faqs/index.md b/topics/galaxy-interface/tutorials/collections/faqs/index.md
index 9ce3fe4fce824b..c66c9e96e0565e 100644
--- a/topics/galaxy-interface/tutorials/collections/faqs/index.md
+++ b/topics/galaxy-interface/tutorials/collections/faqs/index.md
@@ -1,3 +1,5 @@
---
layout: faq-page
+redirect_from:
+- /topics/galaxy-interface/tutorials/processing-many-samples-at-once/faqs/index
---
diff --git a/topics/galaxy-interface/tutorials/processing-many-samples-at-once/faqs/visualisations_igv.md b/topics/galaxy-interface/tutorials/collections/faqs/visualisations_igv.md
similarity index 100%
rename from topics/galaxy-interface/tutorials/processing-many-samples-at-once/faqs/visualisations_igv.md
rename to topics/galaxy-interface/tutorials/collections/faqs/visualisations_igv.md
diff --git a/topics/galaxy-interface/tutorials/collections/tutorial.md b/topics/galaxy-interface/tutorials/collections/tutorial.md
index 552daa1e0b61e0..aa8dab75e312f0 100644
--- a/topics/galaxy-interface/tutorials/collections/tutorial.md
+++ b/topics/galaxy-interface/tutorials/collections/tutorial.md
@@ -2,6 +2,8 @@
layout: tutorial_hands_on
redirect_from:
- /topics/galaxy-data-manipulation/tutorials/collections/tutorial
+ - /topics/galaxy-data-manipulation/tutorials/processing-many-samples-at-once/tutorial
+ - /topics/galaxy-interface/tutorials/processing-many-samples-at-once/tutorial
title: "Using dataset collections"
zenodo_link: "https://doi.org/10.5281/zenodo.5119008"
@@ -46,6 +48,7 @@ recordings:
Here we will show Galaxy features designed to help with the analysis of large numbers of samples. When you have just a few samples - clicking through them is easy. But once you've got hundreds - it becomes very annoying. In Galaxy we have introduced **Dataset collections** that allow you to combine numerous datasets in a single entity that can be easily manipulated.
+
# Getting data
First, we need to upload datasets. Cut and paste the following URLs to Galaxy upload tool (see a {% icon tip %} **Tip** on how to do this [below](#tip-upload-fastqsanger-datasets-via-links)).
diff --git a/topics/galaxy-interface/tutorials/processing-many-samples-at-once/faqs/index.md b/topics/galaxy-interface/tutorials/processing-many-samples-at-once/faqs/index.md
deleted file mode 100644
index 9ce3fe4fce824b..00000000000000
--- a/topics/galaxy-interface/tutorials/processing-many-samples-at-once/faqs/index.md
+++ /dev/null
@@ -1,3 +0,0 @@
----
-layout: faq-page
----
diff --git a/topics/galaxy-interface/tutorials/processing-many-samples-at-once/tutorial.md b/topics/galaxy-interface/tutorials/processing-many-samples-at-once/tutorial.md
deleted file mode 100644
index 5c5d9c2d64a7f1..00000000000000
--- a/topics/galaxy-interface/tutorials/processing-many-samples-at-once/tutorial.md
+++ /dev/null
@@ -1,184 +0,0 @@
----
-layout: tutorial_hands_on
-redirect_from:
- - /topics/galaxy-data-manipulation/tutorials/processing-many-samples-at-once/tutorial
-
-title: "Multisample Analysis"
-tags:
- - collections
-zenodo_link: ""
-level: Advanced
-questions:
-objectives:
-time_estimation: "1h"
-key_points:
-contributors:
- - nekrut
- - pajanne
-subtopic: manage
----
-
-Here we will show Galaxy features designed to help with the analysis of large numbers of samples. When you have just a few samples - clicking through them is easy. But once you've got hundreds - it becomes very annoying. In Galaxy we have introduced **Dataset collections** that allow you to combine numerous datasets in a single entity that can be easily manipulated.
-
-In this tutorial we assume the following:
-
-- you already have basic understanding of how Galaxy works
-- you have an account in Galaxy
-
->
->
-> In this tutorial, we will deal with:
->
-> 1. TOC
-> {:toc}
->
-{: .agenda}
-
-# Getting data
-[In this history](https://test.galaxyproject.org/u/anton/h/collections-1) are a few datasets we will be practicing with (as always with Galaxy tutorial you can upload your own data and play with it instead of the provided datasets):
-
-- `M117-bl_1` - family 117, mother, 1-st (**F**) read from **blood**
-- `M117-bl_2` - family 117, mother, 2-nd (**R**) read from **blood**
-- `M117-ch_1` - family 117, mother, 1-st (**F**) read from **cheek**
-- `M117-ch_1` - family 117, mother, 2-nd (**R**) read from **cheek**
-- `M117C1-bl_1`- family 117, child, 1-st (**F**) read from **blood**
-- `M117C1-bl_2`- family 117, child, 2-nd (**R**) read from **blood**
-- `M117C1-ch_1`- family 117, child, 1-st (**F**) read from **cheek**
-- `M117C1-ch_2`- family 117, child, 2-nd (**R**) read from **cheek**
-
-These datasets represent genomic DNA (enriched for mitochondria via a long range PCR) isolated from blood and cheek (buccal swab) of mother (`M117`) and her child (`M117C1`) that was sequenced on an Illumina miSeq machine as paired-read library (250-bp reads; see our [2014](http://www.pnas.org/content/111/43/15474.abstract) manuscript for **Methods**).
-
-# Creating a list of paired datasets
-
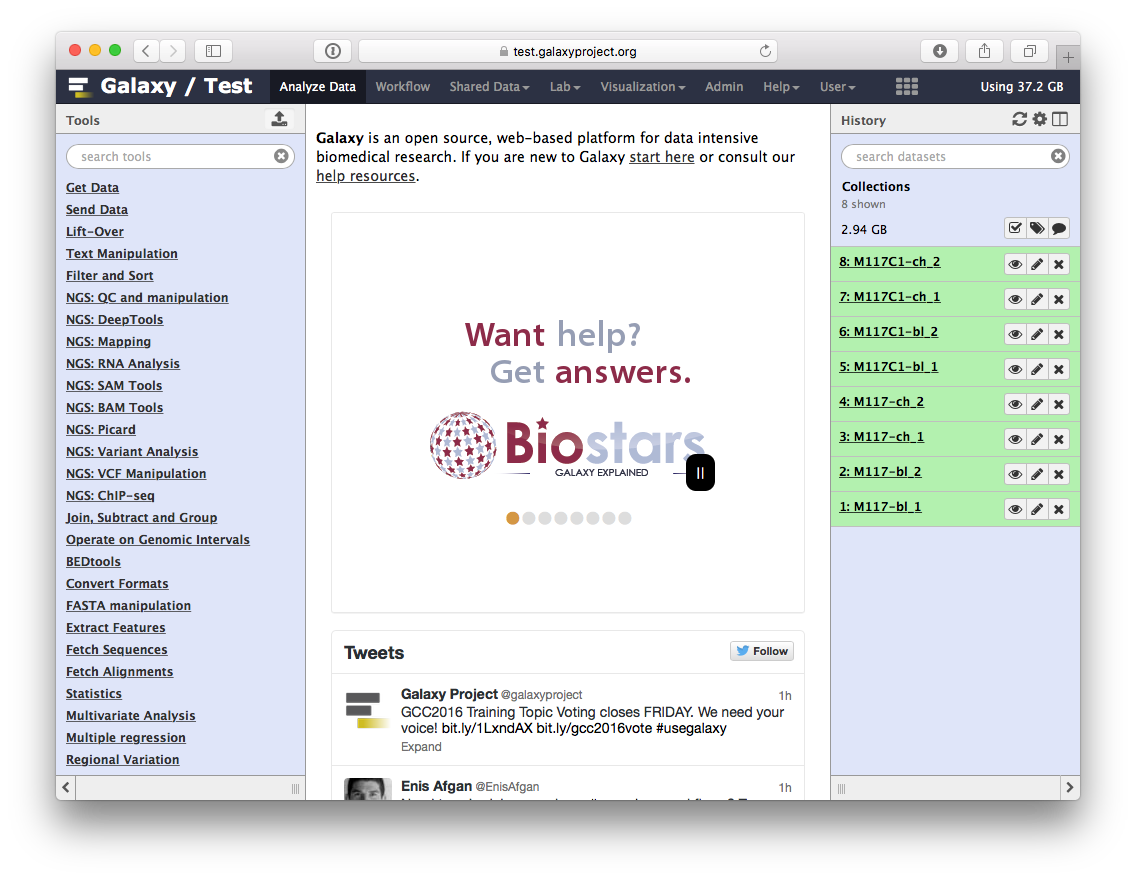
-If you imported [history]( https://test.galaxyproject.org/u/anton/h/collections-1) as described [above](https://github.com/nekrut/galaxy/wiki/Processing-many-samples-at-once#0-getting-data), your screen will look something like this:
-
-
-
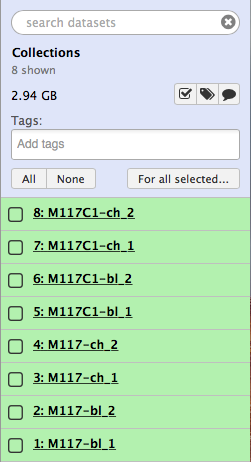
-Now click the checkbox in  and you will see your history changing like this:
-
-
-
-Let's click `All`, which will select all datasets in the history, then click  and finally select **Build List of Dataset Pairs** from the following menu:
-
-
-
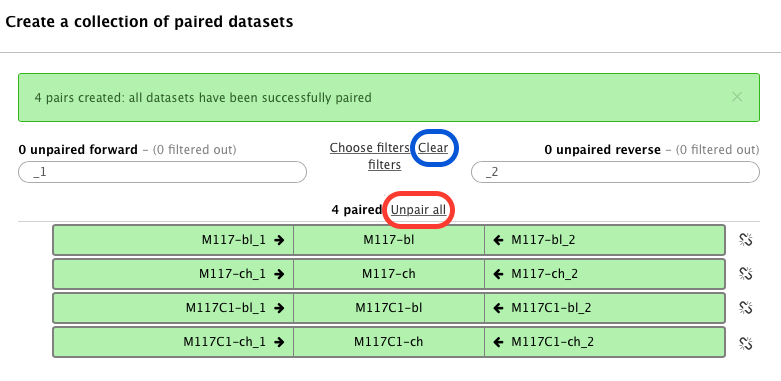
-The following wizard will appear:
-
-
-
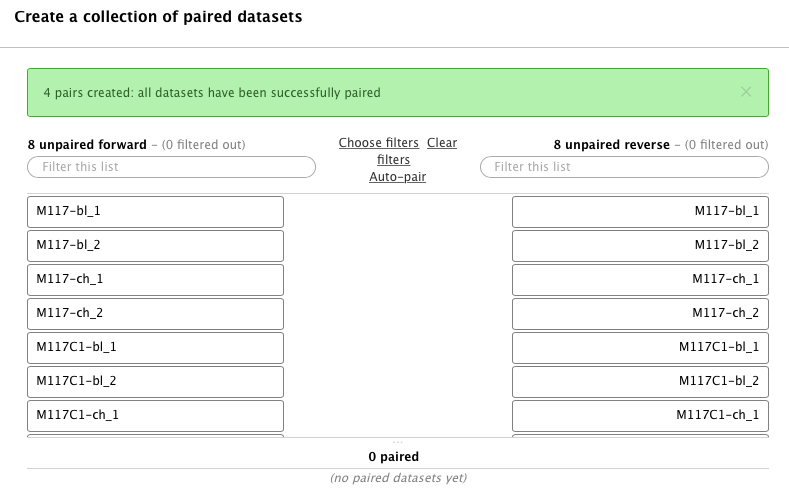
-In this case Galaxy automatically assigned pairs using the `_1` and `_2` endings of dataset names. Let's however pretend that this did not happen. Click on **Unpair all** (highlighted in red in the figure above) link and then on **Clear** link (highlighted in blue in the figure above). The interface will change into its virgin state:
-
-
-
-Hopefully you remember that we have paired-end data in this scenario. Datasets containing the first (forward) and the second (reverse) read are differentiated by having `_1` and `_2` in the filename. We can use this feature in dataset collection wizard to pair our datasets. Type `_1` in the left **Filter this list** text box and `_2` in the right:
-
-
-
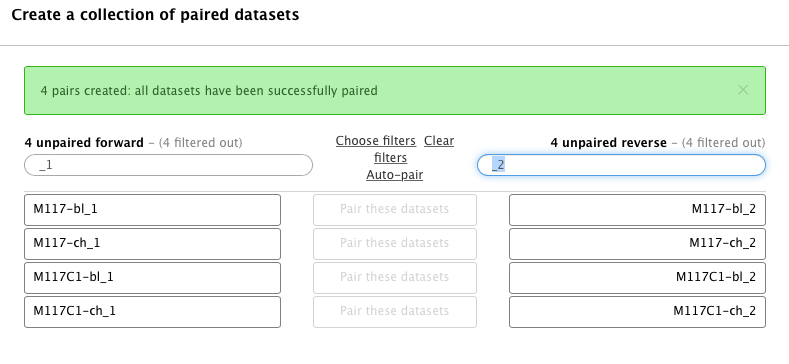
-You will see that the dataset collection wizard will automatically filter lists on each side of the interface:
-
-
-
-Now you can either click **Auto pair** if pairs look good to you (proper combinations of datasets are listed in each line) or pair each forward/reverse group individually by pressing **Pair these datasets** button separating each pair:
-
-
-
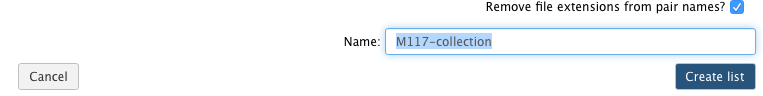
-Now it is time to name the collection:
-
-
-
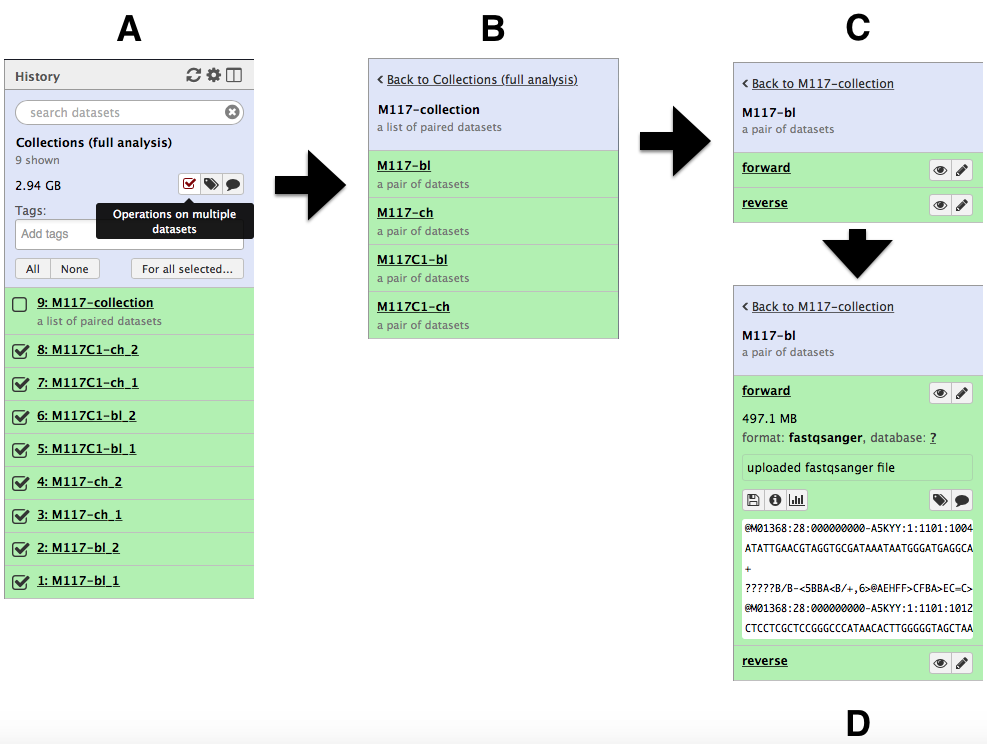
-and create the collection by clicking **Create list**. A new item will appear in the history as you can see on the panel **A** below. Clicking on collection will expand it to show four pairs it contains (panel **B**). Clicking individual pairs will expand them further to reveal **forward** and **reverse** datasets (panel **C**). Expanding these further will enable one to see individual datasets (panel **D**).
-
-
-
-# Using collections
-
-By now we see that a collection can be used to bundle a large number of items into a single history item. This means that many Galaxy tools will be able to process all datasets in a collection transparently to you. Let's try to map these datasets to human genome using `bwa-mem` mapper:
-
-
-
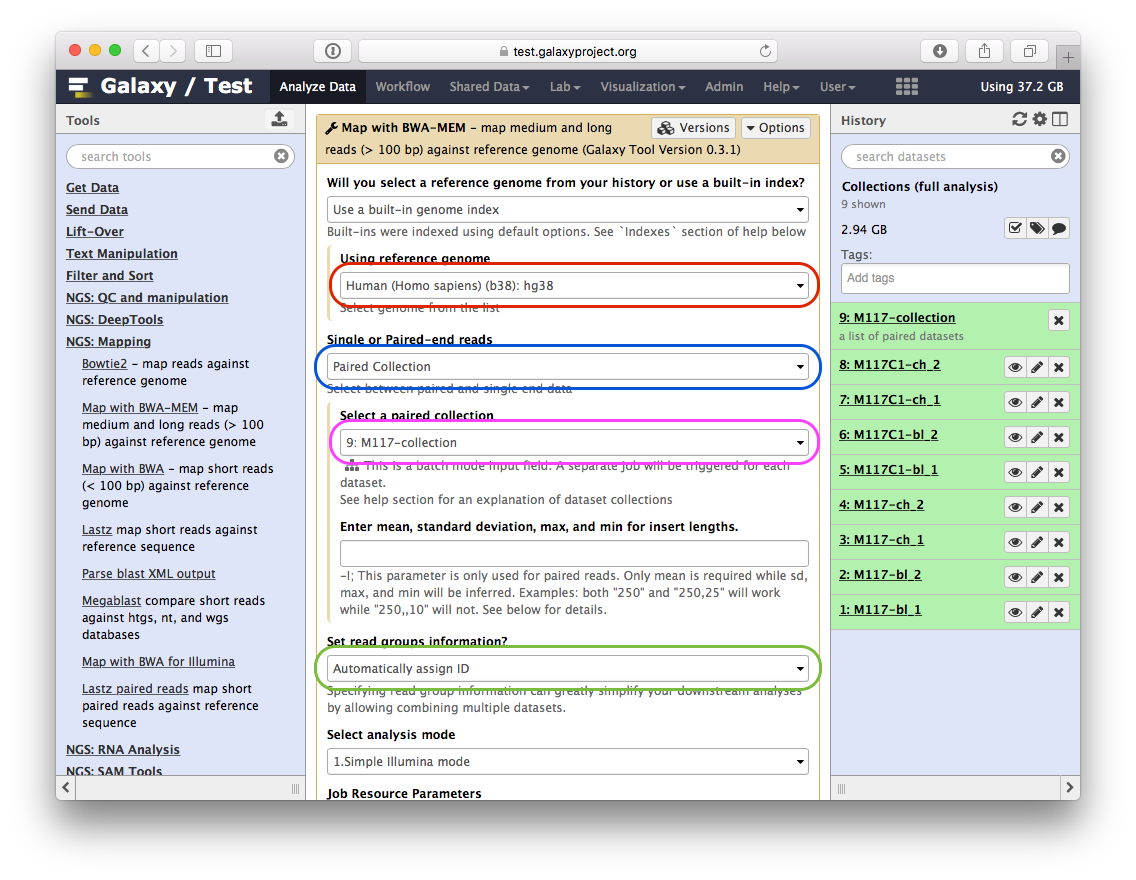
-Here is what you need to do:
-
-- set **Using reference genome** to `hg38` (red outline);
-- set **Single or Paired-end reads** to `Paired collection` (blue outline);
-- select `M177-collection` from **Select a paired collection** dropdown (magenta outline);
-- In **Set read groups information** select `Automatically assign ID` (green outline);
-- scroll down and click **Execute**.
-
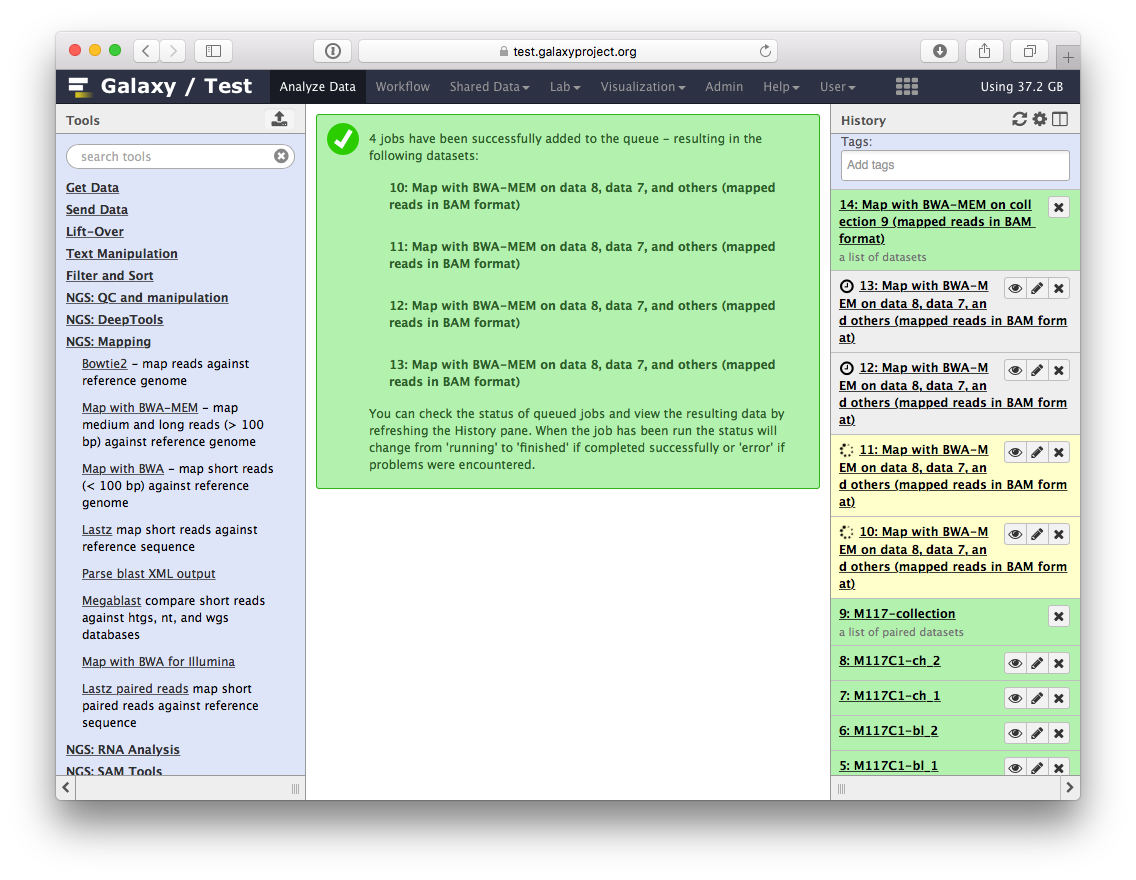
-You will see jobs being submitted and new datasets appearing in the history. IN particular below you can see that Galaxy has started four jobs (two yellow and two gray). This is because we have eight paired datasets with each pair being processed separately by `bwa-mem`. As a result we have four `bwa-mem` runs:
-
-
-
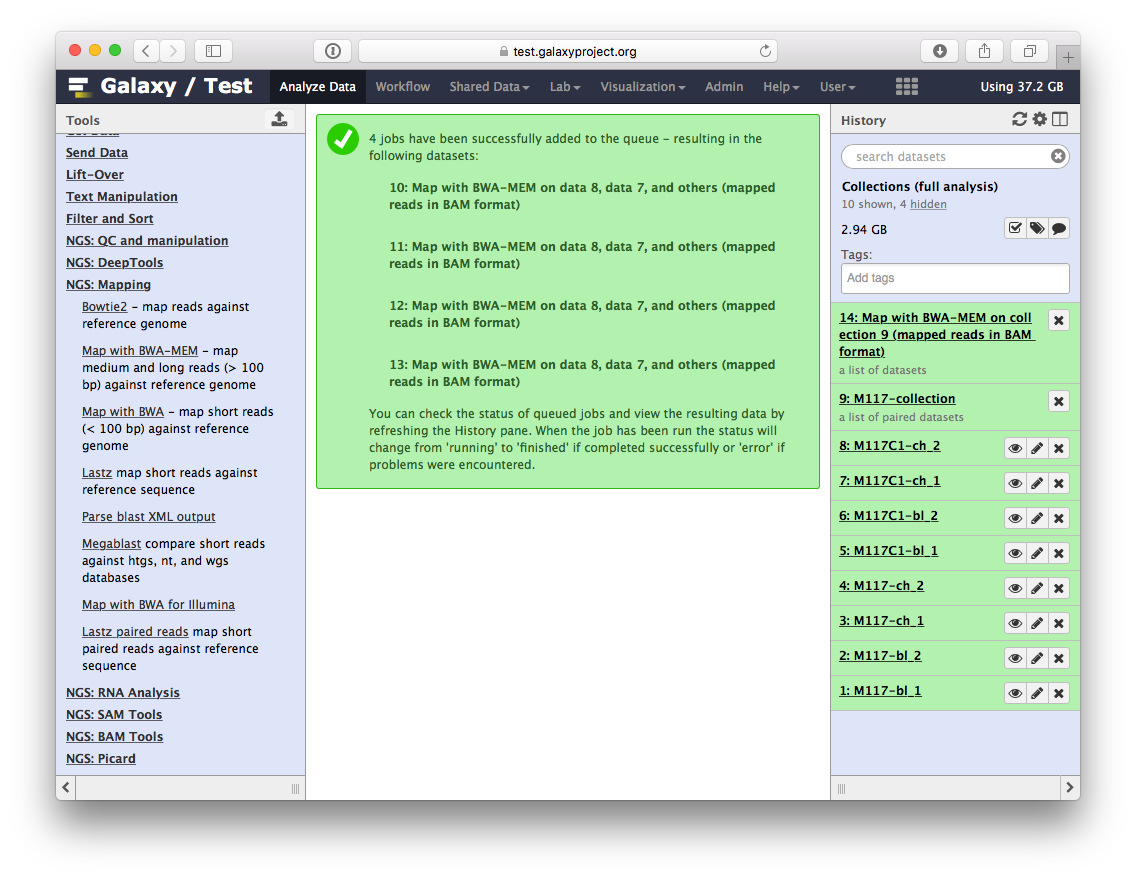
-Once these jobs are finished they will disappear from the history and all results will be represented as a new collection:
-
-
-
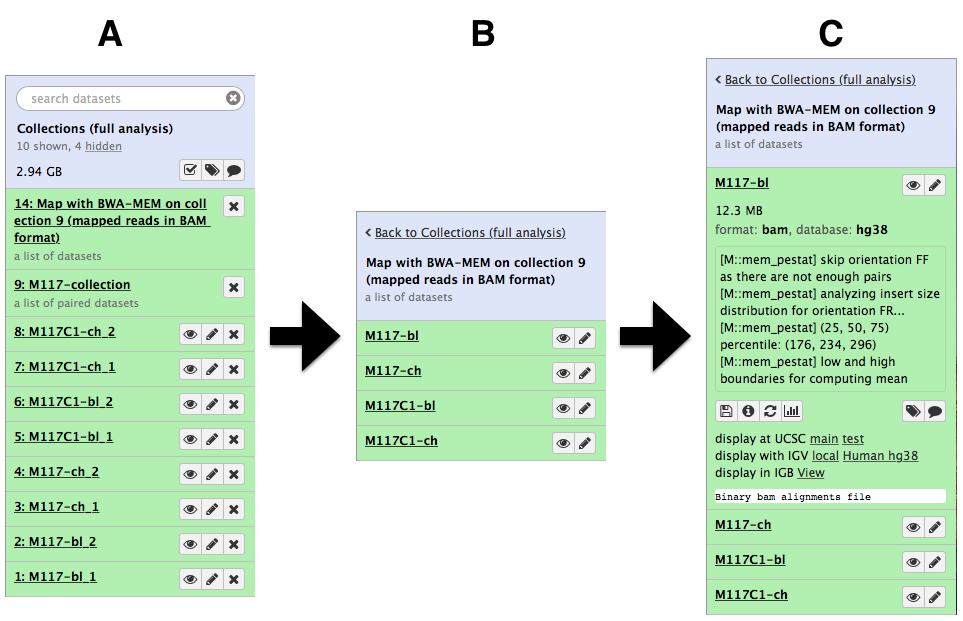
-Let's look at this collection by clicking on it (panel **A** in the figure below). You can see that now this collection is no longer paired (compared to the collection we created in the beginning of this tutorial). This is because `bwa-mem` takes forward and reverse data as input, but produces only a single BAM dataset as the output. So what we have in the result is a *list* of four dataset (BAM files; panels **B** and **C**).
-
-
-
-# Processing collection as a single entity
-
-Now that `bwa-mem` has finished and generated a collection of BAM datasets we can continue to analyze the entire collection as a single Galaxy '*item*'.
-
-## Ensuring consistency of BAM dataset
-
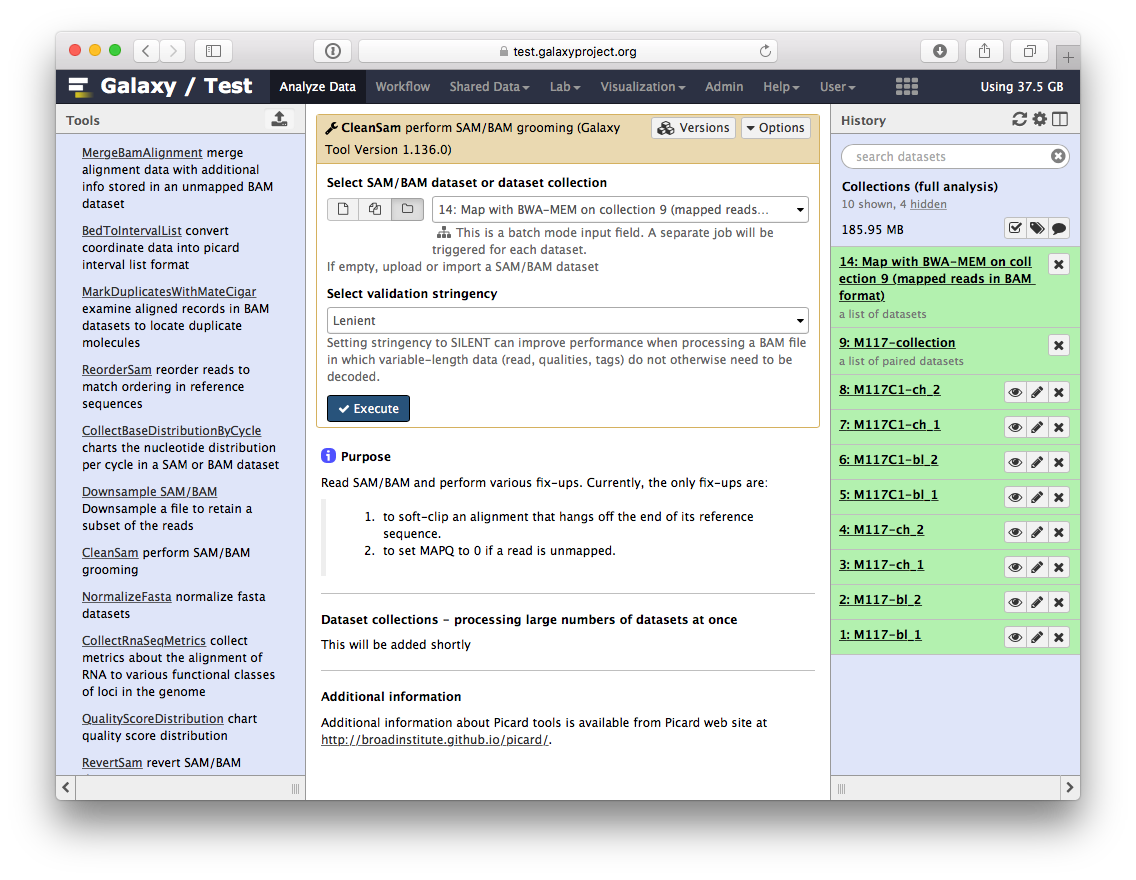
-Let's perform cleanup of our BAM files with `cleanSam` utility from the **Picard** package:
-
-
-
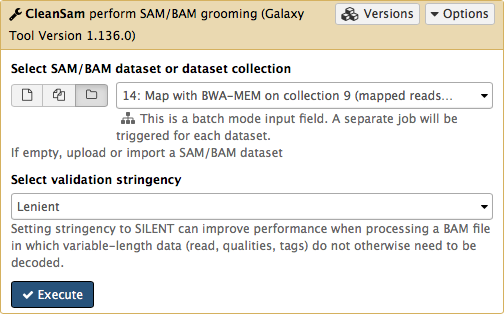
-If you look at the picture above carefully, you will see that the **Select SAM/BAM dataset or dataset collection** parameter is empty (it says `No sam or bam datasets available.`). This is because we do not have single SAM or BAM datasets in the history. Instead we have a collection. So all you need to do is to click on the **folder** () button and you will our BAM collection selected:
-
-
-
-Click **Run Tool**. As an output this tool will produce a collection contained cleaned data.
-
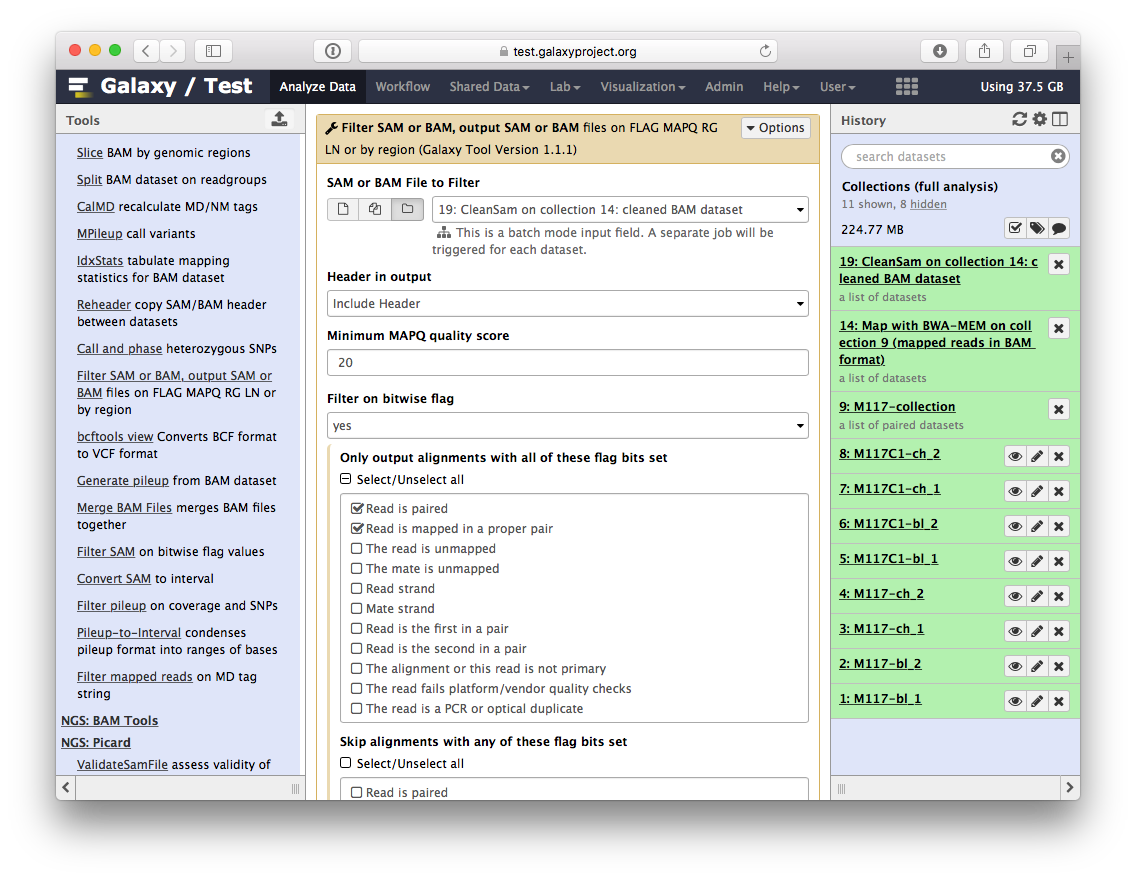
-## Retaining 'proper pairs'
-
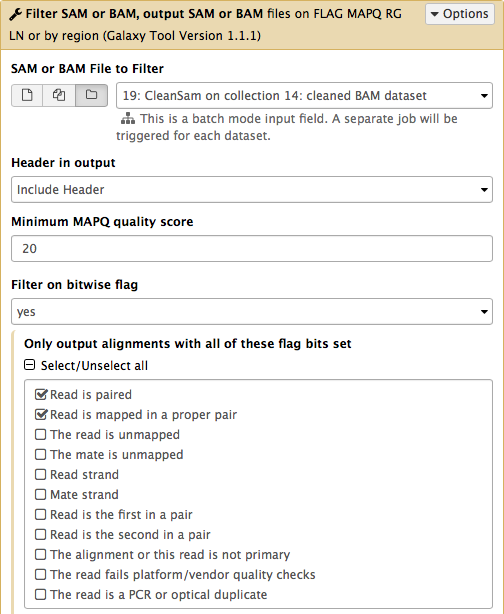
-Now let's clean the dataset further by only preserving truly paired reads (reads satisfying two requirements: (1) read is paired, and (2) it is mapped as a proper pair). For this we will use `Filter SAM or BAM` tools from **SAMTools** collection:
-
-
-
-parameters should be set as shown below. By setting mapping quality to `20` we avoid reads mapping to multiple locations and by using **Filter on bitwise flag** option we ensure that the resulting dataset will contain only properly paired reads. This operation will produce yet another collection containing now filtered datasets.
-
-
-
-## Merging collection into a single dataset
-
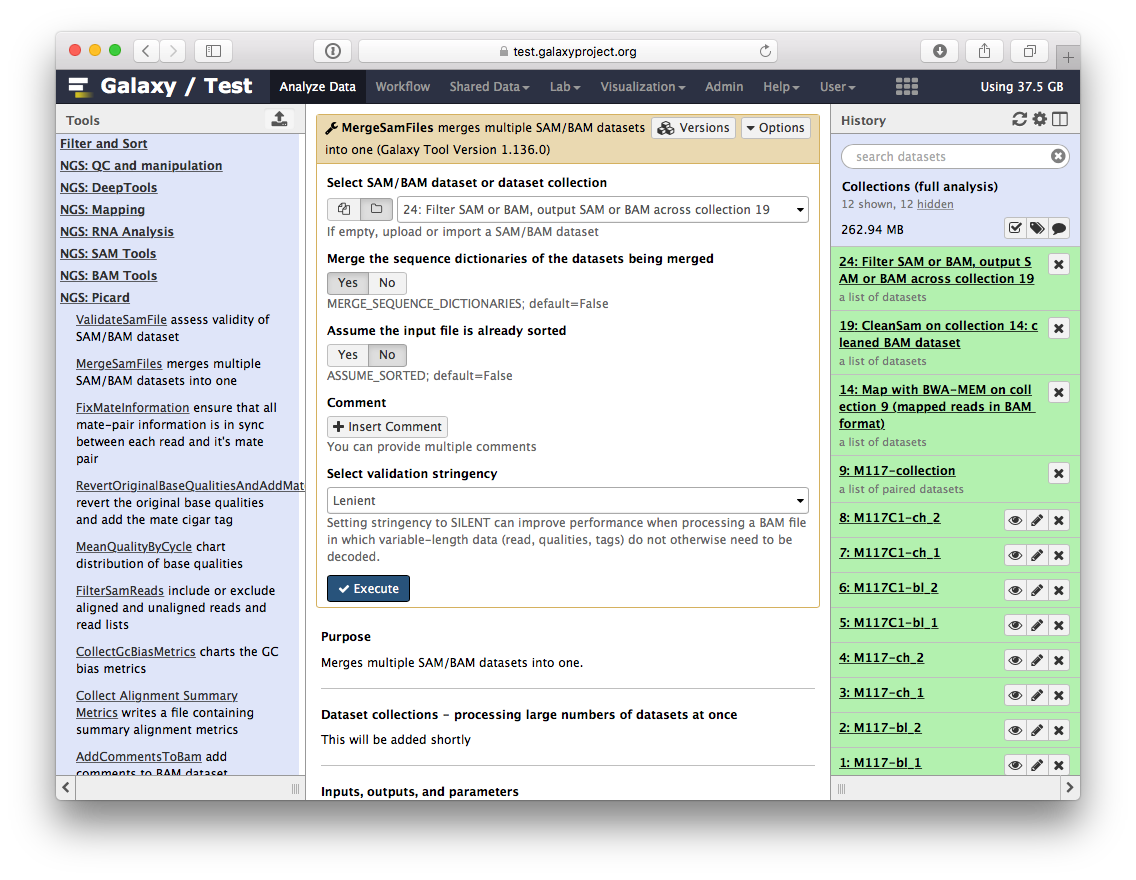
-The beauty of BAM datasets is that they can be combined in a single entity using so called *Read group* ([learn more](https://wiki.galaxyproject.org/Learn/GalaxyNGS101#Understanding_and_manipulating_SAM.2FBAM_datasets) about Read Groups on old wiki, which will be migrated here shortly). This allows to bundle reads from multiple experiments into a single dataset where read identity is maintained by labelling every sequence with *read group* tags. So let's finally reduce this collection to a single BAM dataset. For this we will use `MergeSamFiles` tool for the `Picard` suite:
-
-
-
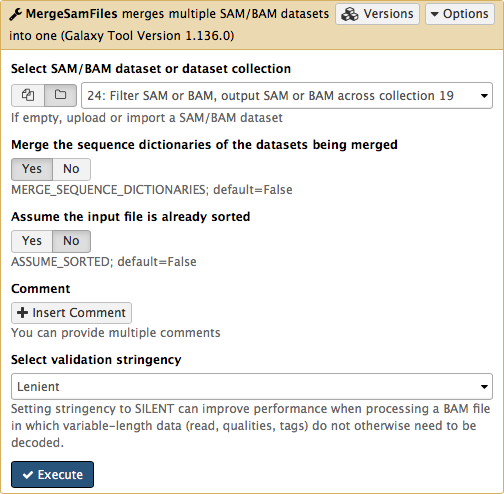
-Here we select the collection generated by the filtering tool described above in [3.1](https://github.com/nekrut/galaxy/wiki/Processing-many-samples-at-once#31-retaining-proper-pairs):
-
-
-
-This operation will **not** generate a collection. Instead, it will generate a single BAM dataset containing mapped reads from our four samples (`M117-bl`, `M117-ch`, `M117C1-bl`, and `M117C1-ch`).
-
-# Let's look at what we've got!
-
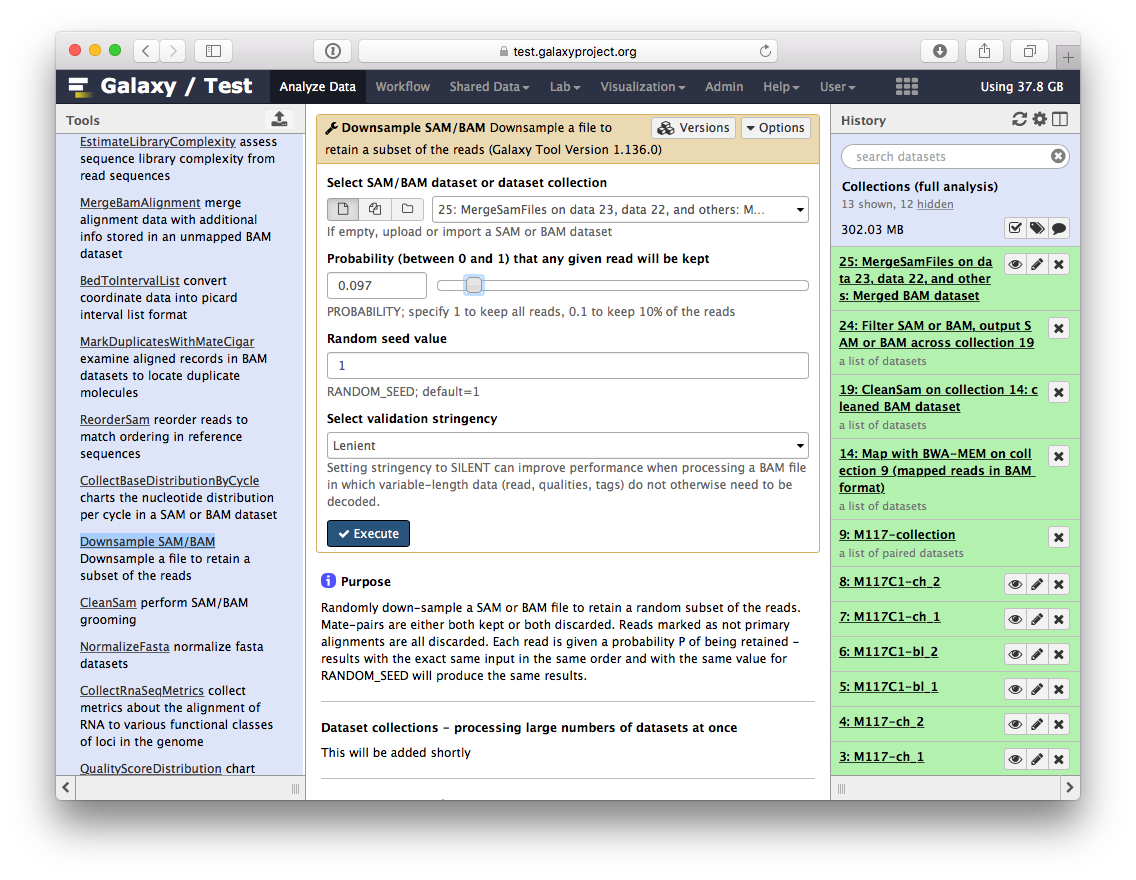
-So we have one BAM dataset combining everything we've done so far. Let's look at the contents of this dataset using a genome browser. First, we will need to downsample the dataset to avoiding overwhelming the browser. For this we will use `Downsample SAM/BAM` tool:
-
-
-
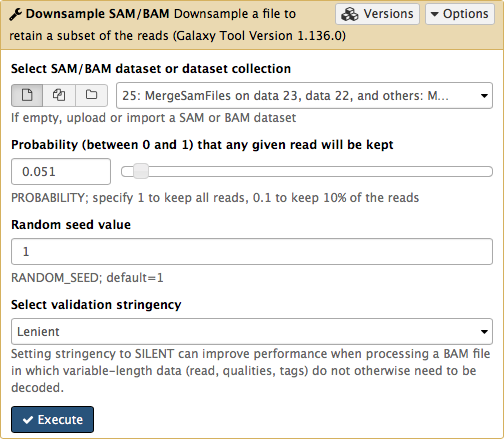
-Set **Probability (between 0 and 1) that any given read will be kept** to roughly `5%` (or `0.05`) using the slider control:
-
-
-
-This will generate another BAM dataset containing only 5% of the original reads and much smaller as a result. Click on this dataset and you will see links to various genome browsers:
-
-
-
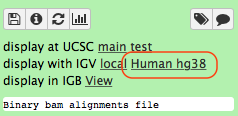
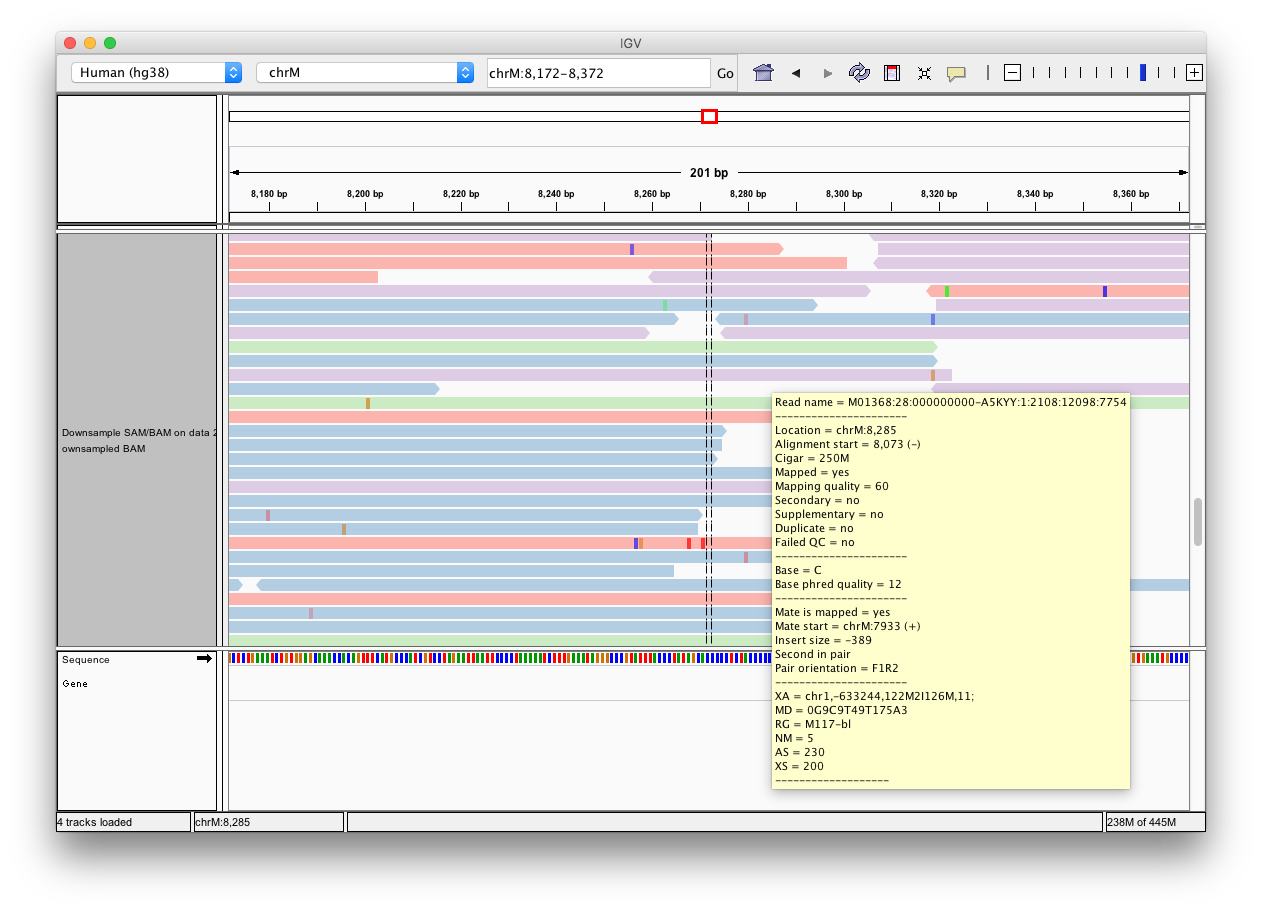
-Click the **Human hg38** link in the **display with IGV** line as highlighted above ([learn](https://wiki.galaxyproject.org/Learn/GalaxyNGS101#Visualizing_multiple_datasets_in_Integrated_Genome_Viewer_.28IGV.29) more about displaying Galaxy data in IGV with this [movie](https://vimeo.com/123442619#t=4m16s)). Below is an example generated with IGV on these data. In this screenshot reads are colored by read group (four distinct colors). A yellow inset displays additional information about a single read. One can see that this read corresponds to read group `M117-bl`.
-
-
-
-# We did not fake this:
-The two histories and the workflow described in this page are accessible directly from this page below:
-
-* History [**Collections**]( https://test.galaxyproject.org/u/anton/h/collections-1)
-* History [**Collections (full analysis)**]( https://test.galaxyproject.org/u/anton/h/collections-full-analysis)
-
-From there you can import histories to make them your own.
-
-# If things don't work...
-...you need to complain. Use [Galaxy's Help Channel](https://help.galaxyproject.org/) to do this.
diff --git a/topics/single-cell/metadata.yaml b/topics/single-cell/metadata.yaml
index b0dadc8ca676e8..7e510aea57ab7b 100644
--- a/topics/single-cell/metadata.yaml
+++ b/topics/single-cell/metadata.yaml
@@ -24,6 +24,9 @@ editorial_board:
- nomadscientist
- mtekman
- pavanvidem
+ - hexhowells
+ - MarisaJL
+ - heylf
toc: true # create table of contents for the subtopics
subtopics:
diff --git a/topics/single-cell/tutorials/scrna-case_FilterPlotandExplore_SeuratTools/tutorial.md b/topics/single-cell/tutorials/scrna-case_FilterPlotandExplore_SeuratTools/tutorial.md
index 79e105e999d852..20e5d614ebf981 100644
--- a/topics/single-cell/tutorials/scrna-case_FilterPlotandExplore_SeuratTools/tutorial.md
+++ b/topics/single-cell/tutorials/scrna-case_FilterPlotandExplore_SeuratTools/tutorial.md
@@ -6,11 +6,6 @@ subtopic: single-cell-CS
priority: 3
zenodo_link: 'https://zenodo.org/record/7053673'
-input_histories:
- - label: "UseGalaxy.eu"
- history: https://singlecell.usegalaxy.eu/u/camila-goclowski/h/tool-based-seurat-fpe-input-data
-
-
questions:
- Is my single cell dataset a quality dataset?
- How do I pick thresholds and parameters in my analysis? What’s a “reasonable” number, and will the world collapse if I pick the wrong one?
diff --git a/topics/single-cell/tutorials/scrna-intro/slides.html b/topics/single-cell/tutorials/scrna-intro/slides.html
index 96448f4d6e0fbf..2aac54b2349798 100644
--- a/topics/single-cell/tutorials/scrna-intro/slides.html
+++ b/topics/single-cell/tutorials/scrna-intro/slides.html
@@ -98,9 +98,9 @@
|Attribute| Summary |
|-:|:-|
-| Resolution| Individual cells within tissues |
-| Signal | Individual gene expression per cell |
-| Differential Expression | Some cells express the same set of genes in the same way; comparing one set of cells against another |
+|Resolution| Individual cells within tissues |
+|Signal | Individual gene expression per cell |
+|Differential Expression | Some cells express the same set of genes in the same way; comparing one set of cells against another |
]
]
@@ -186,7 +186,7 @@
--
-__Biological Replicates__
+
.center[
.reduce90[
diff --git a/topics/single-cell/tutorials/scrna-ncbi-anndata/tutorial.md b/topics/single-cell/tutorials/scrna-ncbi-anndata/tutorial.md
index 09b28413c21803..ff8c939fe52a9c 100644
--- a/topics/single-cell/tutorials/scrna-ncbi-anndata/tutorial.md
+++ b/topics/single-cell/tutorials/scrna-ncbi-anndata/tutorial.md
@@ -230,14 +230,13 @@ The next step is to convert all of the raw files into AnnData objects, this can
{% snippet faqs/galaxy/datasets_add_tag.md %}
> Convert raw data to AnnData
>
-> 1. {% tool [Import AnnData and loom](toolshed.g2.bx.psu.edu/repos/iuc/anndata_import/anndata_import/0.7.5+galaxy1) %} with the following parameters:
-> - *"hd5 format to be created"*: `Anndata file`
-> - *"Format for the annotated data matrix?"*: `Tabular, CSV, TSV`
+> 1. {% tool [Import Anndata](toolshed.g2.bx.psu.edu/repos/iuc/anndata_import/anndata_import/0.10.9+galaxy0) %} with the following parameters:
+> - *"Create anndata from"*: `Tabular, CSV, TSV`
> - *"Annotated data matrix"*
> - {% icon param-files %} *Multiple datasets*: `Select all imported files`
> - *"Does the first column store the row names?"*: `Yes`
>
-> 2. {% tool [Inspect AnnData](toolshed.g2.bx.psu.edu/repos/iuc/anndata_inspect/anndata_inspect/0.7.5+galaxy1) %} with the following parameters:
+> 2. {% tool [Inspect AnnData](toolshed.g2.bx.psu.edu/repos/iuc/anndata_inspect/anndata_inspect/0.10.9+galaxy0) %} with the following parameters:
> - {% icon param-file %} *"Annotated data matrix"*: `select any one of the AnnData files`
> - *"What to inspect?"*: `Key-indexed observations annotation (obs)`
>
@@ -251,7 +250,7 @@ Examine {% icon galaxy-eye %} the {% icon param-file %} *"Inspect AnnData"* outp
> Transpose AnnData objects
>
-> 1. {% tool [Manipulate AnnData](toolshed.g2.bx.psu.edu/repos/iuc/anndata_manipulate/anndata_manipulate/0.7.5+galaxy1) %} with the following parameters:
+> 1. {% tool [Manipulate AnnData](toolshed.g2.bx.psu.edu/repos/iuc/anndata_manipulate/anndata_manipulate/0.10.9+galaxy0) %} with the following parameters:
>
> - *"Annotated data matrix"*
> - {% icon param-files %} *Multiple datasets*: `Select all AnnData files`
@@ -263,7 +262,7 @@ Now, we have all the AnnData objects with the data in the correct orientation. W
> Combine AnnData objects
>
-> 1. {% tool [Manipulate AnnData](toolshed.g2.bx.psu.edu/repos/iuc/anndata_manipulate/anndata_manipulate/0.7.5+galaxy1) %} with the following parameters:
+> 1. {% tool [Manipulate AnnData](toolshed.g2.bx.psu.edu/repos/iuc/anndata_manipulate/anndata_manipulate/0.10.9+galaxy0) %} with the following parameters:
> - {% icon param-file %} *"Annotated data matrix"*: `Select first Manipulate AnnData (transpose) output`
> - *"Function to manipulate the object"*: `Concatenate along the observations axis`
> - {% icon param-file %} *"Annotated data matrix to add"*: `Select all other Manipulate AnnData (transpose) outputs`
@@ -283,7 +282,7 @@ The next step is to annotate our data using the information gathered from the ex
>
>
-> 1. {% tool [Inspect AnnData](toolshed.g2.bx.psu.edu/repos/iuc/anndata_inspect/anndata_inspect/0.7.5+galaxy1) %} with the following parameters:
+> 1. {% tool [Inspect AnnData](toolshed.g2.bx.psu.edu/repos/iuc/anndata_inspect/anndata_inspect/0.10.9+galaxy0) %} with the following parameters:
> - {% icon param-file %} *"Annotated data matrix"*: `Combined Object`
> - *"What to inspect?"*: `Key-indexed observations annotation (obs)`
>
@@ -297,7 +296,7 @@ Let's now add the replicate column which tells us which cells are part of pools
> Create replicate metadata
>
-> 1. {% tool [Replace Text](toolshed.g2.bx.psu.edu/repos/bgruening/text_processing/tp_replace_in_column/1.1.3) %} *in a specific column* with the following parameters:
+> 1. {% tool [Replace Text](toolshed.g2.bx.psu.edu/repos/bgruening/text_processing/tp_replace_in_column/9.3+galaxy1) %} *in a specific column* with the following parameters:
> - {% icon param-file %} *"File to process"*: `Observation data`
> - *"1: Replacement"*
> - *"in column"*: `Column: 2`
@@ -337,7 +336,7 @@ Next we will add the metadata indicating which patient each row came from.
> Create patient data
>
-> 1. {% tool [Replace Text](toolshed.g2.bx.psu.edu/repos/bgruening/text_processing/tp_replace_in_column/1.1.3) %} with the following parameters:
+> 1. {% tool [Replace Text](toolshed.g2.bx.psu.edu/repos/bgruening/text_processing/tp_replace_in_column/9.3+galaxy1) %} with the following parameters:
> - {% icon param-file %} *"File to process"*: `Observation data`
> - *"1: Replacement"*
> - *"in column"*: `Column: 2`
@@ -372,7 +371,7 @@ We will now add a column to indicate which sample each row came from using the s
> Create sample ID metadata
>
-> 1. {% tool [Replace Text](toolshed.g2.bx.psu.edu/repos/bgruening/text_processing/tp_replace_in_column/1.1.3) %} with the following parameters:
+> 1. {% tool [Replace Text](toolshed.g2.bx.psu.edu/repos/bgruening/text_processing/tp_replace_in_column/9.3+galaxy1) %} with the following parameters:
> - {% icon param-file %} *"File to process"*: `Observation data`
> - *"1: Replacement"*
> - *"in column"*: `Column: 2`
@@ -428,7 +427,7 @@ Finally we will add the tumor column which indicates which tumor sample each row
> Create tumor metadata
>
-> 1. {% tool [Replace Text](toolshed.g2.bx.psu.edu/repos/bgruening/text_processing/tp_replace_in_column/1.1.3) %} with the following parameters:
+> 1. {% tool [Replace Text](toolshed.g2.bx.psu.edu/repos/bgruening/text_processing/tp_replace_in_column/9.3+galaxy1) %} with the following parameters:
> - {% icon param-file %} *"File to process"*: `Observation data`
> - *"1: Replacement"*
> - *"in column"*: `Column: 2`
@@ -495,7 +494,7 @@ With the metadata table ready, we can add it to our original combined object!
> Add metadata to AnnData object
>
-> 1. {% tool [Manipulate AnnData](toolshed.g2.bx.psu.edu/repos/iuc/anndata_manipulate/anndata_manipulate/0.7.5+galaxy1) %} with the following parameters:
+> 1. {% tool [Manipulate AnnData](toolshed.g2.bx.psu.edu/repos/iuc/anndata_manipulate/anndata_manipulate/0.10.9+galaxy0) %} with the following parameters:
> - {% icon param-file %} *"Annotated data matrix"*: `Combined Object`
> - *"Function to manipulate the object"*: `Add new annotation(s) for observations of variables`
> - *"What to annotate?"*: `Observations (obs)`
@@ -513,13 +512,9 @@ With the manual annotations added, we need to do some further processing to add
First, we will run the ```Scanpy FilterCells``` tool without actually filtering. This tool will add some metadata about the counts and numbers of expressed genes.
-> Scanpy FilterCells version issue
-> The {% tool [Scanpy FilterCells](toolshed.g2.bx.psu.edu/repos/ebi-gxa/scanpy_filter_cells/scanpy_filter_cells/1.8.1+galaxy9) %} tool (used below) does not work in the latest version (**1.8.1+galaxy93**), switch to version **1.8.1+galaxy9** in order to run the tool without error.
-{: .warning}
-
> Add initial metadata
>
-> 1. {% tool [Scanpy FilterCells](toolshed.g2.bx.psu.edu/repos/ebi-gxa/scanpy_filter_cells/scanpy_filter_cells/1.8.1+galaxy9) %} with the following parameters:
+> 1. {% tool [Scanpy FilterCells](toolshed.g2.bx.psu.edu/repos/ebi-gxa/scanpy_filter_cells/scanpy_filter_cells/1.9.3+galaxy0) %} with the following parameters:
> - {% icon param-file %} *"Input object in AnnData/Loom format"*: `Annotated Object`
> - *"Name of the column in `anndata.var` that contains gene name"*: `_index`
>
@@ -529,7 +524,7 @@ The final tool, {% icon tool %} **AnnData Operations**, will add the rest of our
> Add final metadata
>
-> 1. {% tool [AnnData Operations](toolshed.g2.bx.psu.edu/repos/ebi-gxa/anndata_ops/anndata_ops/1.8.1+galaxy9) %} with the following parameters:
+> 1. {% tool [AnnData Operations](toolshed.g2.bx.psu.edu/repos/ebi-gxa/anndata_ops/anndata_ops/1.9.3+galaxy0) %} with the following parameters:
> - {% icon param-file %} *"Input object in hdf5 AnnData format"*: `Output of Scanpy FilterCells`
> - **+ Insert Flag genes that start with these names**
> - *"1: Parameters to select cells to keep"*
@@ -538,7 +533,7 @@ The final tool, {% icon tool %} **AnnData Operations**, will add the rest of our
>
> 2. **Rename** {% icon galaxy-pencil %} output `Final Object`
>
-> 3. {% tool [Inspect AnnData](toolshed.g2.bx.psu.edu/repos/iuc/anndata_inspect/anndata_inspect/0.7.5+galaxy1) %} with the following parameters:
+> 3. {% tool [Inspect AnnData](toolshed.g2.bx.psu.edu/repos/iuc/anndata_inspect/anndata_inspect/0.10.9+galaxy0) %} with the following parameters:
> - {% icon param-file %} *"Annotated data matrix"*: `Final Object`
> - *"What to inspect?"*: `Key-indexed observations annotation (obs)`
>
diff --git a/topics/single-cell/tutorials/scrna-ncbi-anndata/workflows/NCBI_to_Anndata.ga b/topics/single-cell/tutorials/scrna-ncbi-anndata/workflows/NCBI_to_Anndata.ga
index 44b91122375eaf..9130363e6fc0b6 100644
--- a/topics/single-cell/tutorials/scrna-ncbi-anndata/workflows/NCBI_to_Anndata.ga
+++ b/topics/single-cell/tutorials/scrna-ncbi-anndata/workflows/NCBI_to_Anndata.ga
@@ -1,6 +1,99 @@
{
"a_galaxy_workflow": "true",
"annotation": "",
+ "comments": [
+ {
+ "child_steps": [
+ 30,
+ 29,
+ 19,
+ 20,
+ 10,
+ 21,
+ 11,
+ 22,
+ 12,
+ 23,
+ 13,
+ 24,
+ 14,
+ 25,
+ 15,
+ 26,
+ 16,
+ 27,
+ 17,
+ 28,
+ 18
+ ],
+ "color": "blue",
+ "data": {
+ "title": "Creating the AnnData object"
+ },
+ "id": 0,
+ "position": [
+ 258.9,
+ 0
+ ],
+ "size": [
+ 811.1,
+ 1584.5
+ ],
+ "type": "frame"
+ },
+ {
+ "child_steps": [
+ 45,
+ 46,
+ 44
+ ],
+ "color": "lime",
+ "data": {
+ "title": "Adding quality control metrics"
+ },
+ "id": 2,
+ "position": [
+ 3070,
+ 634
+ ],
+ "size": [
+ 800,
+ 206.5
+ ],
+ "type": "frame"
+ },
+ {
+ "child_steps": [
+ 38,
+ 31,
+ 36,
+ 37,
+ 39,
+ 42,
+ 32,
+ 33,
+ 34,
+ 35,
+ 40,
+ 41,
+ 43
+ ],
+ "color": "orange",
+ "data": {
+ "title": "Annotation the data"
+ },
+ "id": 1,
+ "position": [
+ 1110,
+ 219.4
+ ],
+ "size": [
+ 1920,
+ 614.5
+ ],
+ "type": "frame"
+ }
+ ],
"creator": [
{
"class": "Person",
@@ -16,6 +109,9 @@
"format-version": "0.1",
"license": "CC-BY-4.0",
"name": "NCBI to Anndata",
+ "report": {
+ "markdown": "\n# Workflow Execution Report\n\n## Workflow Inputs\n```galaxy\ninvocation_inputs()\n```\n\n## Workflow Outputs\n```galaxy\ninvocation_outputs()\n```\n\n## Workflow\n```galaxy\nworkflow_display()\n```\n"
+ },
"steps": {
"0": {
"annotation": "",
@@ -34,7 +130,7 @@
"outputs": [],
"position": {
"left": 0,
- "top": 6
+ "top": 71.39838831245815
},
"tool_id": null,
"tool_state": "{\"optional\": false, \"tag\": null}",
@@ -61,7 +157,7 @@
"outputs": [],
"position": {
"left": 0,
- "top": 160
+ "top": 225.39838831245814
},
"tool_id": null,
"tool_state": "{\"optional\": false, \"tag\": null}",
@@ -88,7 +184,7 @@
"outputs": [],
"position": {
"left": 0,
- "top": 314
+ "top": 379.39838831245817
},
"tool_id": null,
"tool_state": "{\"optional\": false, \"tag\": null}",
@@ -115,7 +211,7 @@
"outputs": [],
"position": {
"left": 0,
- "top": 468
+ "top": 533.3983883124581
},
"tool_id": null,
"tool_state": "{\"optional\": false, \"tag\": null}",
@@ -142,7 +238,7 @@
"outputs": [],
"position": {
"left": 0,
- "top": 622
+ "top": 687.3983883124581
},
"tool_id": null,
"tool_state": "{\"optional\": false, \"tag\": null}",
@@ -169,7 +265,7 @@
"outputs": [],
"position": {
"left": 0,
- "top": 776
+ "top": 841.3983883124581
},
"tool_id": null,
"tool_state": "{\"optional\": false, \"tag\": null}",
@@ -196,7 +292,7 @@
"outputs": [],
"position": {
"left": 0,
- "top": 930
+ "top": 995.3983883124581
},
"tool_id": null,
"tool_state": "{\"optional\": false, \"tag\": null}",
@@ -223,7 +319,7 @@
"outputs": [],
"position": {
"left": 0,
- "top": 1084
+ "top": 1149.398388312458
},
"tool_id": null,
"tool_state": "{\"optional\": false, \"tag\": null}",
@@ -250,7 +346,7 @@
"outputs": [],
"position": {
"left": 0,
- "top": 1238
+ "top": 1303.398388312458
},
"tool_id": null,
"tool_state": "{\"optional\": false, \"tag\": null}",
@@ -277,7 +373,7 @@
"outputs": [],
"position": {
"left": 0,
- "top": 1392
+ "top": 1457.398388312458
},
"tool_id": null,
"tool_state": "{\"optional\": false, \"tag\": null}",
@@ -289,18 +385,23 @@
},
"10": {
"annotation": "",
- "content_id": "toolshed.g2.bx.psu.edu/repos/iuc/anndata_import/anndata_import/0.7.5+galaxy1",
+ "content_id": "toolshed.g2.bx.psu.edu/repos/iuc/anndata_import/anndata_import/0.10.9+galaxy0",
"errors": null,
"id": 10,
"input_connections": {
- "hd5_format|in|input": {
+ "in|input": {
"id": 0,
"output_name": "output"
}
},
- "inputs": [],
+ "inputs": [
+ {
+ "description": "runtime parameter for tool Import Anndata",
+ "name": "in"
+ }
+ ],
"label": null,
- "name": "Import Anndata and loom",
+ "name": "Import Anndata",
"outputs": [
{
"name": "anndata",
@@ -308,19 +409,19 @@
}
],
"position": {
- "left": 279.9999176480769,
- "top": 0
+ "left": 279.97219340870714,
+ "top": 40
},
"post_job_actions": {},
- "tool_id": "toolshed.g2.bx.psu.edu/repos/iuc/anndata_import/anndata_import/0.7.5+galaxy1",
+ "tool_id": "toolshed.g2.bx.psu.edu/repos/iuc/anndata_import/anndata_import/0.10.9+galaxy0",
"tool_shed_repository": {
- "changeset_revision": "fb9e030ffae4",
+ "changeset_revision": "6f143dfe902a",
"name": "anndata_import",
"owner": "iuc",
"tool_shed": "toolshed.g2.bx.psu.edu"
},
- "tool_state": "{\"__input_ext\": \"input\", \"chromInfo\": \"/opt/galaxy/tool-data/shared/ucsc/chrom/?.len\", \"hd5_format\": {\"filetype\": \"anndata\", \"__current_case__\": 0, \"in\": {\"adata_format\": \"tabular\", \"__current_case__\": 1, \"input\": {\"__class__\": \"ConnectedValue\"}, \"first_column_names\": true}}, \"__page__\": null, \"__rerun_remap_job_id__\": null}",
- "tool_version": "0.7.5+galaxy1",
+ "tool_state": "{\"in\": {\"adata_format\": \"tabular\", \"__current_case__\": 1, \"input\": {\"__class__\": \"ConnectedValue\"}, \"first_column_names\": true}, \"__page__\": null, \"__rerun_remap_job_id__\": null}",
+ "tool_version": "0.10.9+galaxy0",
"type": "tool",
"uuid": "09d068b4-2600-40ca-9d30-216b6568b0aa",
"when": null,
@@ -328,18 +429,23 @@
},
"11": {
"annotation": "",
- "content_id": "toolshed.g2.bx.psu.edu/repos/iuc/anndata_import/anndata_import/0.7.5+galaxy1",
+ "content_id": "toolshed.g2.bx.psu.edu/repos/iuc/anndata_import/anndata_import/0.10.9+galaxy0",
"errors": null,
"id": 11,
"input_connections": {
- "hd5_format|in|input": {
+ "in|input": {
"id": 1,
"output_name": "output"
}
},
- "inputs": [],
+ "inputs": [
+ {
+ "description": "runtime parameter for tool Import Anndata",
+ "name": "in"
+ }
+ ],
"label": null,
- "name": "Import Anndata and loom",
+ "name": "Import Anndata",
"outputs": [
{
"name": "anndata",
@@ -347,19 +453,19 @@
}
],
"position": {
- "left": 279.9999176480769,
- "top": 154
+ "left": 279.018522318486,
+ "top": 218.4261525685412
},
"post_job_actions": {},
- "tool_id": "toolshed.g2.bx.psu.edu/repos/iuc/anndata_import/anndata_import/0.7.5+galaxy1",
+ "tool_id": "toolshed.g2.bx.psu.edu/repos/iuc/anndata_import/anndata_import/0.10.9+galaxy0",
"tool_shed_repository": {
- "changeset_revision": "fb9e030ffae4",
+ "changeset_revision": "6f143dfe902a",
"name": "anndata_import",
"owner": "iuc",
"tool_shed": "toolshed.g2.bx.psu.edu"
},
- "tool_state": "{\"__input_ext\": \"input\", \"chromInfo\": \"/opt/galaxy/tool-data/shared/ucsc/chrom/?.len\", \"hd5_format\": {\"filetype\": \"anndata\", \"__current_case__\": 0, \"in\": {\"adata_format\": \"tabular\", \"__current_case__\": 1, \"input\": {\"__class__\": \"ConnectedValue\"}, \"first_column_names\": true}}, \"__page__\": null, \"__rerun_remap_job_id__\": null}",
- "tool_version": "0.7.5+galaxy1",
+ "tool_state": "{\"in\": {\"adata_format\": \"tabular\", \"__current_case__\": 1, \"input\": {\"__class__\": \"ConnectedValue\"}, \"first_column_names\": true}, \"__page__\": null, \"__rerun_remap_job_id__\": null}",
+ "tool_version": "0.10.9+galaxy0",
"type": "tool",
"uuid": "b69371fd-59c6-4ea7-ba48-bac55f945bad",
"when": null,
@@ -367,18 +473,23 @@
},
"12": {
"annotation": "",
- "content_id": "toolshed.g2.bx.psu.edu/repos/iuc/anndata_import/anndata_import/0.7.5+galaxy1",
+ "content_id": "toolshed.g2.bx.psu.edu/repos/iuc/anndata_import/anndata_import/0.10.9+galaxy0",
"errors": null,
"id": 12,
"input_connections": {
- "hd5_format|in|input": {
+ "in|input": {
"id": 2,
"output_name": "output"
}
},
- "inputs": [],
+ "inputs": [
+ {
+ "description": "runtime parameter for tool Import Anndata",
+ "name": "in"
+ }
+ ],
"label": null,
- "name": "Import Anndata and loom",
+ "name": "Import Anndata",
"outputs": [
{
"name": "anndata",
@@ -386,19 +497,19 @@
}
],
"position": {
- "left": 279.9999176480769,
- "top": 308
+ "left": 278.8572183008857,
+ "top": 372.26958955669994
},
"post_job_actions": {},
- "tool_id": "toolshed.g2.bx.psu.edu/repos/iuc/anndata_import/anndata_import/0.7.5+galaxy1",
+ "tool_id": "toolshed.g2.bx.psu.edu/repos/iuc/anndata_import/anndata_import/0.10.9+galaxy0",
"tool_shed_repository": {
- "changeset_revision": "fb9e030ffae4",
+ "changeset_revision": "6f143dfe902a",
"name": "anndata_import",
"owner": "iuc",
"tool_shed": "toolshed.g2.bx.psu.edu"
},
- "tool_state": "{\"__input_ext\": \"input\", \"chromInfo\": \"/opt/galaxy/tool-data/shared/ucsc/chrom/?.len\", \"hd5_format\": {\"filetype\": \"anndata\", \"__current_case__\": 0, \"in\": {\"adata_format\": \"tabular\", \"__current_case__\": 1, \"input\": {\"__class__\": \"ConnectedValue\"}, \"first_column_names\": true}}, \"__page__\": null, \"__rerun_remap_job_id__\": null}",
- "tool_version": "0.7.5+galaxy1",
+ "tool_state": "{\"in\": {\"adata_format\": \"tabular\", \"__current_case__\": 1, \"input\": {\"__class__\": \"ConnectedValue\"}, \"first_column_names\": true}, \"__page__\": null, \"__rerun_remap_job_id__\": null}",
+ "tool_version": "0.10.9+galaxy0",
"type": "tool",
"uuid": "99a950da-eb84-45c4-b4b6-8d0216940828",
"when": null,
@@ -406,18 +517,23 @@
},
"13": {
"annotation": "",
- "content_id": "toolshed.g2.bx.psu.edu/repos/iuc/anndata_import/anndata_import/0.7.5+galaxy1",
+ "content_id": "toolshed.g2.bx.psu.edu/repos/iuc/anndata_import/anndata_import/0.10.9+galaxy0",
"errors": null,
"id": 13,
"input_connections": {
- "hd5_format|in|input": {
+ "in|input": {
"id": 3,
"output_name": "output"
}
},
- "inputs": [],
+ "inputs": [
+ {
+ "description": "runtime parameter for tool Import Anndata",
+ "name": "in"
+ }
+ ],
"label": null,
- "name": "Import Anndata and loom",
+ "name": "Import Anndata",
"outputs": [
{
"name": "anndata",
@@ -426,18 +542,18 @@
],
"position": {
"left": 279.9999176480769,
- "top": 462
+ "top": 527.3983883124581
},
"post_job_actions": {},
- "tool_id": "toolshed.g2.bx.psu.edu/repos/iuc/anndata_import/anndata_import/0.7.5+galaxy1",
+ "tool_id": "toolshed.g2.bx.psu.edu/repos/iuc/anndata_import/anndata_import/0.10.9+galaxy0",
"tool_shed_repository": {
- "changeset_revision": "fb9e030ffae4",
+ "changeset_revision": "6f143dfe902a",
"name": "anndata_import",
"owner": "iuc",
"tool_shed": "toolshed.g2.bx.psu.edu"
},
- "tool_state": "{\"__input_ext\": \"input\", \"chromInfo\": \"/opt/galaxy/tool-data/shared/ucsc/chrom/?.len\", \"hd5_format\": {\"filetype\": \"anndata\", \"__current_case__\": 0, \"in\": {\"adata_format\": \"tabular\", \"__current_case__\": 1, \"input\": {\"__class__\": \"ConnectedValue\"}, \"first_column_names\": true}}, \"__page__\": null, \"__rerun_remap_job_id__\": null}",
- "tool_version": "0.7.5+galaxy1",
+ "tool_state": "{\"in\": {\"adata_format\": \"tabular\", \"__current_case__\": 1, \"input\": {\"__class__\": \"ConnectedValue\"}, \"first_column_names\": true}, \"__page__\": null, \"__rerun_remap_job_id__\": null}",
+ "tool_version": "0.10.9+galaxy0",
"type": "tool",
"uuid": "fc0c681e-1aac-40ae-8298-aabc01e5a87b",
"when": null,
@@ -445,18 +561,23 @@
},
"14": {
"annotation": "",
- "content_id": "toolshed.g2.bx.psu.edu/repos/iuc/anndata_import/anndata_import/0.7.5+galaxy1",
+ "content_id": "toolshed.g2.bx.psu.edu/repos/iuc/anndata_import/anndata_import/0.10.9+galaxy0",
"errors": null,
"id": 14,
"input_connections": {
- "hd5_format|in|input": {
+ "in|input": {
"id": 4,
"output_name": "output"
}
},
- "inputs": [],
+ "inputs": [
+ {
+ "description": "runtime parameter for tool Import Anndata",
+ "name": "in"
+ }
+ ],
"label": null,
- "name": "Import Anndata and loom",
+ "name": "Import Anndata",
"outputs": [
{
"name": "anndata",
@@ -464,19 +585,19 @@
}
],
"position": {
- "left": 279.9999176480769,
- "top": 616
+ "left": 278.8572958107027,
+ "top": 681.3983593877771
},
"post_job_actions": {},
- "tool_id": "toolshed.g2.bx.psu.edu/repos/iuc/anndata_import/anndata_import/0.7.5+galaxy1",
+ "tool_id": "toolshed.g2.bx.psu.edu/repos/iuc/anndata_import/anndata_import/0.10.9+galaxy0",
"tool_shed_repository": {
- "changeset_revision": "fb9e030ffae4",
+ "changeset_revision": "6f143dfe902a",
"name": "anndata_import",
"owner": "iuc",
"tool_shed": "toolshed.g2.bx.psu.edu"
},
- "tool_state": "{\"__input_ext\": \"input\", \"chromInfo\": \"/opt/galaxy/tool-data/shared/ucsc/chrom/?.len\", \"hd5_format\": {\"filetype\": \"anndata\", \"__current_case__\": 0, \"in\": {\"adata_format\": \"tabular\", \"__current_case__\": 1, \"input\": {\"__class__\": \"ConnectedValue\"}, \"first_column_names\": true}}, \"__page__\": null, \"__rerun_remap_job_id__\": null}",
- "tool_version": "0.7.5+galaxy1",
+ "tool_state": "{\"in\": {\"adata_format\": \"tabular\", \"__current_case__\": 1, \"input\": {\"__class__\": \"ConnectedValue\"}, \"first_column_names\": true}, \"__page__\": null, \"__rerun_remap_job_id__\": null}",
+ "tool_version": "0.10.9+galaxy0",
"type": "tool",
"uuid": "b5c7249c-c358-4a71-a637-12f269e009ab",
"when": null,
@@ -484,18 +605,23 @@
},
"15": {
"annotation": "",
- "content_id": "toolshed.g2.bx.psu.edu/repos/iuc/anndata_import/anndata_import/0.7.5+galaxy1",
+ "content_id": "toolshed.g2.bx.psu.edu/repos/iuc/anndata_import/anndata_import/0.10.9+galaxy0",
"errors": null,
"id": 15,
"input_connections": {
- "hd5_format|in|input": {
+ "in|input": {
"id": 5,
"output_name": "output"
}
},
- "inputs": [],
+ "inputs": [
+ {
+ "description": "runtime parameter for tool Import Anndata",
+ "name": "in"
+ }
+ ],
"label": null,
- "name": "Import Anndata and loom",
+ "name": "Import Anndata",
"outputs": [
{
"name": "anndata",
@@ -504,18 +630,18 @@
],
"position": {
"left": 279.9999176480769,
- "top": 770
+ "top": 835.3983883124581
},
"post_job_actions": {},
- "tool_id": "toolshed.g2.bx.psu.edu/repos/iuc/anndata_import/anndata_import/0.7.5+galaxy1",
+ "tool_id": "toolshed.g2.bx.psu.edu/repos/iuc/anndata_import/anndata_import/0.10.9+galaxy0",
"tool_shed_repository": {
- "changeset_revision": "fb9e030ffae4",
+ "changeset_revision": "6f143dfe902a",
"name": "anndata_import",
"owner": "iuc",
"tool_shed": "toolshed.g2.bx.psu.edu"
},
- "tool_state": "{\"__input_ext\": \"input\", \"chromInfo\": \"/opt/galaxy/tool-data/shared/ucsc/chrom/?.len\", \"hd5_format\": {\"filetype\": \"anndata\", \"__current_case__\": 0, \"in\": {\"adata_format\": \"tabular\", \"__current_case__\": 1, \"input\": {\"__class__\": \"ConnectedValue\"}, \"first_column_names\": true}}, \"__page__\": null, \"__rerun_remap_job_id__\": null}",
- "tool_version": "0.7.5+galaxy1",
+ "tool_state": "{\"in\": {\"adata_format\": \"tabular\", \"__current_case__\": 1, \"input\": {\"__class__\": \"ConnectedValue\"}, \"first_column_names\": true}, \"__page__\": null, \"__rerun_remap_job_id__\": null}",
+ "tool_version": "0.10.9+galaxy0",
"type": "tool",
"uuid": "bbc79158-5e04-49db-8088-37da78a2c83f",
"when": null,
@@ -523,18 +649,23 @@
},
"16": {
"annotation": "",
- "content_id": "toolshed.g2.bx.psu.edu/repos/iuc/anndata_import/anndata_import/0.7.5+galaxy1",
+ "content_id": "toolshed.g2.bx.psu.edu/repos/iuc/anndata_import/anndata_import/0.10.9+galaxy0",
"errors": null,
"id": 16,
"input_connections": {
- "hd5_format|in|input": {
+ "in|input": {
"id": 6,
"output_name": "output"
}
},
- "inputs": [],
+ "inputs": [
+ {
+ "description": "runtime parameter for tool Import Anndata",
+ "name": "in"
+ }
+ ],
"label": null,
- "name": "Import Anndata and loom",
+ "name": "Import Anndata",
"outputs": [
{
"name": "anndata",
@@ -542,19 +673,19 @@
}
],
"position": {
- "left": 279.9999176480769,
- "top": 924
+ "left": 279.9861240698978,
+ "top": 988.0815036417088
},
"post_job_actions": {},
- "tool_id": "toolshed.g2.bx.psu.edu/repos/iuc/anndata_import/anndata_import/0.7.5+galaxy1",
+ "tool_id": "toolshed.g2.bx.psu.edu/repos/iuc/anndata_import/anndata_import/0.10.9+galaxy0",
"tool_shed_repository": {
- "changeset_revision": "fb9e030ffae4",
+ "changeset_revision": "6f143dfe902a",
"name": "anndata_import",
"owner": "iuc",
"tool_shed": "toolshed.g2.bx.psu.edu"
},
- "tool_state": "{\"__input_ext\": \"input\", \"chromInfo\": \"/opt/galaxy/tool-data/shared/ucsc/chrom/?.len\", \"hd5_format\": {\"filetype\": \"anndata\", \"__current_case__\": 0, \"in\": {\"adata_format\": \"tabular\", \"__current_case__\": 1, \"input\": {\"__class__\": \"ConnectedValue\"}, \"first_column_names\": true}}, \"__page__\": null, \"__rerun_remap_job_id__\": null}",
- "tool_version": "0.7.5+galaxy1",
+ "tool_state": "{\"in\": {\"adata_format\": \"tabular\", \"__current_case__\": 1, \"input\": {\"__class__\": \"ConnectedValue\"}, \"first_column_names\": true}, \"__page__\": null, \"__rerun_remap_job_id__\": null}",
+ "tool_version": "0.10.9+galaxy0",
"type": "tool",
"uuid": "177a94fa-ea39-4e04-9d95-47d84bee200d",
"when": null,
@@ -562,18 +693,23 @@
},
"17": {
"annotation": "",
- "content_id": "toolshed.g2.bx.psu.edu/repos/iuc/anndata_import/anndata_import/0.7.5+galaxy1",
+ "content_id": "toolshed.g2.bx.psu.edu/repos/iuc/anndata_import/anndata_import/0.10.9+galaxy0",
"errors": null,
"id": 17,
"input_connections": {
- "hd5_format|in|input": {
+ "in|input": {
"id": 7,
"output_name": "output"
}
},
- "inputs": [],
+ "inputs": [
+ {
+ "description": "runtime parameter for tool Import Anndata",
+ "name": "in"
+ }
+ ],
"label": null,
- "name": "Import Anndata and loom",
+ "name": "Import Anndata",
"outputs": [
{
"name": "anndata",
@@ -581,19 +717,19 @@
}
],
"position": {
- "left": 279.9999176480769,
- "top": 1078
+ "left": 279.9861487853543,
+ "top": 1143.3983751589587
},
"post_job_actions": {},
- "tool_id": "toolshed.g2.bx.psu.edu/repos/iuc/anndata_import/anndata_import/0.7.5+galaxy1",
+ "tool_id": "toolshed.g2.bx.psu.edu/repos/iuc/anndata_import/anndata_import/0.10.9+galaxy0",
"tool_shed_repository": {
- "changeset_revision": "fb9e030ffae4",
+ "changeset_revision": "6f143dfe902a",
"name": "anndata_import",
"owner": "iuc",
"tool_shed": "toolshed.g2.bx.psu.edu"
},
- "tool_state": "{\"__input_ext\": \"input\", \"chromInfo\": \"/opt/galaxy/tool-data/shared/ucsc/chrom/?.len\", \"hd5_format\": {\"filetype\": \"anndata\", \"__current_case__\": 0, \"in\": {\"adata_format\": \"tabular\", \"__current_case__\": 1, \"input\": {\"__class__\": \"ConnectedValue\"}, \"first_column_names\": true}}, \"__page__\": null, \"__rerun_remap_job_id__\": null}",
- "tool_version": "0.7.5+galaxy1",
+ "tool_state": "{\"in\": {\"adata_format\": \"tabular\", \"__current_case__\": 1, \"input\": {\"__class__\": \"ConnectedValue\"}, \"first_column_names\": true}, \"__page__\": null, \"__rerun_remap_job_id__\": null}",
+ "tool_version": "0.10.9+galaxy0",
"type": "tool",
"uuid": "3a263cfa-a15c-4565-b2cd-49989db25371",
"when": null,
@@ -601,18 +737,23 @@
},
"18": {
"annotation": "",
- "content_id": "toolshed.g2.bx.psu.edu/repos/iuc/anndata_import/anndata_import/0.7.5+galaxy1",
+ "content_id": "toolshed.g2.bx.psu.edu/repos/iuc/anndata_import/anndata_import/0.10.9+galaxy0",
"errors": null,
"id": 18,
"input_connections": {
- "hd5_format|in|input": {
+ "in|input": {
"id": 8,
"output_name": "output"
}
},
- "inputs": [],
+ "inputs": [
+ {
+ "description": "runtime parameter for tool Import Anndata",
+ "name": "in"
+ }
+ ],
"label": null,
- "name": "Import Anndata and loom",
+ "name": "Import Anndata",
"outputs": [
{
"name": "anndata",
@@ -621,18 +762,18 @@
],
"position": {
"left": 279.9999176480769,
- "top": 1232
+ "top": 1297.398388312458
},
"post_job_actions": {},
- "tool_id": "toolshed.g2.bx.psu.edu/repos/iuc/anndata_import/anndata_import/0.7.5+galaxy1",
+ "tool_id": "toolshed.g2.bx.psu.edu/repos/iuc/anndata_import/anndata_import/0.10.9+galaxy0",
"tool_shed_repository": {
- "changeset_revision": "fb9e030ffae4",
+ "changeset_revision": "6f143dfe902a",
"name": "anndata_import",
"owner": "iuc",
"tool_shed": "toolshed.g2.bx.psu.edu"
},
- "tool_state": "{\"__input_ext\": \"input\", \"chromInfo\": \"/opt/galaxy/tool-data/shared/ucsc/chrom/?.len\", \"hd5_format\": {\"filetype\": \"anndata\", \"__current_case__\": 0, \"in\": {\"adata_format\": \"tabular\", \"__current_case__\": 1, \"input\": {\"__class__\": \"ConnectedValue\"}, \"first_column_names\": true}}, \"__page__\": null, \"__rerun_remap_job_id__\": null}",
- "tool_version": "0.7.5+galaxy1",
+ "tool_state": "{\"in\": {\"adata_format\": \"tabular\", \"__current_case__\": 1, \"input\": {\"__class__\": \"ConnectedValue\"}, \"first_column_names\": true}, \"__page__\": null, \"__rerun_remap_job_id__\": null}",
+ "tool_version": "0.10.9+galaxy0",
"type": "tool",
"uuid": "2a7d63ed-774f-4894-af10-92e06131adcf",
"when": null,
@@ -640,18 +781,23 @@
},
"19": {
"annotation": "",
- "content_id": "toolshed.g2.bx.psu.edu/repos/iuc/anndata_import/anndata_import/0.7.5+galaxy1",
+ "content_id": "toolshed.g2.bx.psu.edu/repos/iuc/anndata_import/anndata_import/0.10.9+galaxy0",
"errors": null,
"id": 19,
"input_connections": {
- "hd5_format|in|input": {
+ "in|input": {
"id": 9,
"output_name": "output"
}
},
- "inputs": [],
+ "inputs": [
+ {
+ "description": "runtime parameter for tool Import Anndata",
+ "name": "in"
+ }
+ ],
"label": null,
- "name": "Import Anndata and loom",
+ "name": "Import Anndata",
"outputs": [
{
"name": "anndata",
@@ -660,18 +806,18 @@
],
"position": {
"left": 279.9999176480769,
- "top": 1386
+ "top": 1451.398388312458
},
"post_job_actions": {},
- "tool_id": "toolshed.g2.bx.psu.edu/repos/iuc/anndata_import/anndata_import/0.7.5+galaxy1",
+ "tool_id": "toolshed.g2.bx.psu.edu/repos/iuc/anndata_import/anndata_import/0.10.9+galaxy0",
"tool_shed_repository": {
- "changeset_revision": "fb9e030ffae4",
+ "changeset_revision": "6f143dfe902a",
"name": "anndata_import",
"owner": "iuc",
"tool_shed": "toolshed.g2.bx.psu.edu"
},
- "tool_state": "{\"__input_ext\": \"input\", \"chromInfo\": \"/opt/galaxy/tool-data/shared/ucsc/chrom/?.len\", \"hd5_format\": {\"filetype\": \"anndata\", \"__current_case__\": 0, \"in\": {\"adata_format\": \"tabular\", \"__current_case__\": 1, \"input\": {\"__class__\": \"ConnectedValue\"}, \"first_column_names\": true}}, \"__page__\": null, \"__rerun_remap_job_id__\": null}",
- "tool_version": "0.7.5+galaxy1",
+ "tool_state": "{\"in\": {\"adata_format\": \"tabular\", \"__current_case__\": 1, \"input\": {\"__class__\": \"ConnectedValue\"}, \"first_column_names\": true}, \"__page__\": null, \"__rerun_remap_job_id__\": null}",
+ "tool_version": "0.10.9+galaxy0",
"type": "tool",
"uuid": "51f452a2-b1a5-4315-bb26-0093c79a3aa9",
"when": null,
@@ -679,7 +825,7 @@
},
"20": {
"annotation": "",
- "content_id": "toolshed.g2.bx.psu.edu/repos/iuc/anndata_manipulate/anndata_manipulate/0.7.5+galaxy1",
+ "content_id": "toolshed.g2.bx.psu.edu/repos/iuc/anndata_manipulate/anndata_manipulate/0.10.9+galaxy0",
"errors": null,
"id": 20,
"input_connections": {
@@ -698,19 +844,19 @@
}
],
"position": {
- "left": 559.9999540284473,
- "top": 0
+ "left": 559.986130131553,
+ "top": 64.08156964137
},
"post_job_actions": {},
- "tool_id": "toolshed.g2.bx.psu.edu/repos/iuc/anndata_manipulate/anndata_manipulate/0.7.5+galaxy1",
+ "tool_id": "toolshed.g2.bx.psu.edu/repos/iuc/anndata_manipulate/anndata_manipulate/0.10.9+galaxy0",
"tool_shed_repository": {
- "changeset_revision": "3d748954434b",
+ "changeset_revision": "c4209ea387d4",
"name": "anndata_manipulate",
"owner": "iuc",
"tool_shed": "toolshed.g2.bx.psu.edu"
},
- "tool_state": "{\"__input_ext\": \"h5ad\", \"chromInfo\": \"/opt/galaxy/tool-data/shared/ucsc/chrom/?.len\", \"input\": {\"__class__\": \"ConnectedValue\"}, \"manipulate\": {\"function\": \"transpose\", \"__current_case__\": 5}, \"__page__\": null, \"__rerun_remap_job_id__\": null}",
- "tool_version": "0.7.5+galaxy1",
+ "tool_state": "{\"input\": {\"__class__\": \"ConnectedValue\"}, \"manipulate\": {\"function\": \"transpose\", \"__current_case__\": 7}, \"__page__\": null, \"__rerun_remap_job_id__\": null}",
+ "tool_version": "0.10.9+galaxy0",
"type": "tool",
"uuid": "defbd9c2-d44c-48c8-8ed6-b6649dcae1af",
"when": null,
@@ -718,7 +864,7 @@
},
"21": {
"annotation": "",
- "content_id": "toolshed.g2.bx.psu.edu/repos/iuc/anndata_manipulate/anndata_manipulate/0.7.5+galaxy1",
+ "content_id": "toolshed.g2.bx.psu.edu/repos/iuc/anndata_manipulate/anndata_manipulate/0.10.9+galaxy0",
"errors": null,
"id": 21,
"input_connections": {
@@ -738,18 +884,18 @@
],
"position": {
"left": 559.9999540284473,
- "top": 154
+ "top": 219.39838831245814
},
"post_job_actions": {},
- "tool_id": "toolshed.g2.bx.psu.edu/repos/iuc/anndata_manipulate/anndata_manipulate/0.7.5+galaxy1",
+ "tool_id": "toolshed.g2.bx.psu.edu/repos/iuc/anndata_manipulate/anndata_manipulate/0.10.9+galaxy0",
"tool_shed_repository": {
- "changeset_revision": "3d748954434b",
+ "changeset_revision": "c4209ea387d4",
"name": "anndata_manipulate",
"owner": "iuc",
"tool_shed": "toolshed.g2.bx.psu.edu"
},
- "tool_state": "{\"__input_ext\": \"h5ad\", \"chromInfo\": \"/opt/galaxy/tool-data/shared/ucsc/chrom/?.len\", \"input\": {\"__class__\": \"ConnectedValue\"}, \"manipulate\": {\"function\": \"transpose\", \"__current_case__\": 5}, \"__page__\": null, \"__rerun_remap_job_id__\": null}",
- "tool_version": "0.7.5+galaxy1",
+ "tool_state": "{\"input\": {\"__class__\": \"ConnectedValue\"}, \"manipulate\": {\"function\": \"transpose\", \"__current_case__\": 7}, \"__page__\": null, \"__rerun_remap_job_id__\": null}",
+ "tool_version": "0.10.9+galaxy0",
"type": "tool",
"uuid": "a6c1c73b-1c2a-421b-ae4c-d79e286614ea",
"when": null,
@@ -757,7 +903,7 @@
},
"22": {
"annotation": "",
- "content_id": "toolshed.g2.bx.psu.edu/repos/iuc/anndata_manipulate/anndata_manipulate/0.7.5+galaxy1",
+ "content_id": "toolshed.g2.bx.psu.edu/repos/iuc/anndata_manipulate/anndata_manipulate/0.10.9+galaxy0",
"errors": null,
"id": 22,
"input_connections": {
@@ -777,18 +923,18 @@
],
"position": {
"left": 559.9999540284473,
- "top": 308
+ "top": 373.39838831245817
},
"post_job_actions": {},
- "tool_id": "toolshed.g2.bx.psu.edu/repos/iuc/anndata_manipulate/anndata_manipulate/0.7.5+galaxy1",
+ "tool_id": "toolshed.g2.bx.psu.edu/repos/iuc/anndata_manipulate/anndata_manipulate/0.10.9+galaxy0",
"tool_shed_repository": {
- "changeset_revision": "3d748954434b",
+ "changeset_revision": "c4209ea387d4",
"name": "anndata_manipulate",
"owner": "iuc",
"tool_shed": "toolshed.g2.bx.psu.edu"
},
- "tool_state": "{\"__input_ext\": \"h5ad\", \"chromInfo\": \"/opt/galaxy/tool-data/shared/ucsc/chrom/?.len\", \"input\": {\"__class__\": \"ConnectedValue\"}, \"manipulate\": {\"function\": \"transpose\", \"__current_case__\": 5}, \"__page__\": null, \"__rerun_remap_job_id__\": null}",
- "tool_version": "0.7.5+galaxy1",
+ "tool_state": "{\"input\": {\"__class__\": \"ConnectedValue\"}, \"manipulate\": {\"function\": \"transpose\", \"__current_case__\": 7}, \"__page__\": null, \"__rerun_remap_job_id__\": null}",
+ "tool_version": "0.10.9+galaxy0",
"type": "tool",
"uuid": "fd73dc52-957e-433a-9aab-733efb9df757",
"when": null,
@@ -796,7 +942,7 @@
},
"23": {
"annotation": "",
- "content_id": "toolshed.g2.bx.psu.edu/repos/iuc/anndata_manipulate/anndata_manipulate/0.7.5+galaxy1",
+ "content_id": "toolshed.g2.bx.psu.edu/repos/iuc/anndata_manipulate/anndata_manipulate/0.10.9+galaxy0",
"errors": null,
"id": 23,
"input_connections": {
@@ -816,18 +962,18 @@
],
"position": {
"left": 559.9999540284473,
- "top": 462
+ "top": 527.3983883124581
},
"post_job_actions": {},
- "tool_id": "toolshed.g2.bx.psu.edu/repos/iuc/anndata_manipulate/anndata_manipulate/0.7.5+galaxy1",
+ "tool_id": "toolshed.g2.bx.psu.edu/repos/iuc/anndata_manipulate/anndata_manipulate/0.10.9+galaxy0",
"tool_shed_repository": {
- "changeset_revision": "3d748954434b",
+ "changeset_revision": "c4209ea387d4",
"name": "anndata_manipulate",
"owner": "iuc",
"tool_shed": "toolshed.g2.bx.psu.edu"
},
- "tool_state": "{\"__input_ext\": \"h5ad\", \"chromInfo\": \"/opt/galaxy/tool-data/shared/ucsc/chrom/?.len\", \"input\": {\"__class__\": \"ConnectedValue\"}, \"manipulate\": {\"function\": \"transpose\", \"__current_case__\": 5}, \"__page__\": null, \"__rerun_remap_job_id__\": null}",
- "tool_version": "0.7.5+galaxy1",
+ "tool_state": "{\"input\": {\"__class__\": \"ConnectedValue\"}, \"manipulate\": {\"function\": \"transpose\", \"__current_case__\": 7}, \"__page__\": null, \"__rerun_remap_job_id__\": null}",
+ "tool_version": "0.10.9+galaxy0",
"type": "tool",
"uuid": "fe89b9a5-dd11-4750-bdff-998cd62b8c76",
"when": null,
@@ -835,7 +981,7 @@
},
"24": {
"annotation": "",
- "content_id": "toolshed.g2.bx.psu.edu/repos/iuc/anndata_manipulate/anndata_manipulate/0.7.5+galaxy1",
+ "content_id": "toolshed.g2.bx.psu.edu/repos/iuc/anndata_manipulate/anndata_manipulate/0.10.9+galaxy0",
"errors": null,
"id": 24,
"input_connections": {
@@ -855,18 +1001,18 @@
],
"position": {
"left": 559.9999540284473,
- "top": 616
+ "top": 681.3983883124581
},
"post_job_actions": {},
- "tool_id": "toolshed.g2.bx.psu.edu/repos/iuc/anndata_manipulate/anndata_manipulate/0.7.5+galaxy1",
+ "tool_id": "toolshed.g2.bx.psu.edu/repos/iuc/anndata_manipulate/anndata_manipulate/0.10.9+galaxy0",
"tool_shed_repository": {
- "changeset_revision": "3d748954434b",
+ "changeset_revision": "c4209ea387d4",
"name": "anndata_manipulate",
"owner": "iuc",
"tool_shed": "toolshed.g2.bx.psu.edu"
},
- "tool_state": "{\"__input_ext\": \"h5ad\", \"chromInfo\": \"/opt/galaxy/tool-data/shared/ucsc/chrom/?.len\", \"input\": {\"__class__\": \"ConnectedValue\"}, \"manipulate\": {\"function\": \"transpose\", \"__current_case__\": 5}, \"__page__\": null, \"__rerun_remap_job_id__\": null}",
- "tool_version": "0.7.5+galaxy1",
+ "tool_state": "{\"input\": {\"__class__\": \"ConnectedValue\"}, \"manipulate\": {\"function\": \"transpose\", \"__current_case__\": 7}, \"__page__\": null, \"__rerun_remap_job_id__\": null}",
+ "tool_version": "0.10.9+galaxy0",
"type": "tool",
"uuid": "75fd4890-4bce-4978-a636-f3ec85e3af63",
"when": null,
@@ -874,7 +1020,7 @@
},
"25": {
"annotation": "",
- "content_id": "toolshed.g2.bx.psu.edu/repos/iuc/anndata_manipulate/anndata_manipulate/0.7.5+galaxy1",
+ "content_id": "toolshed.g2.bx.psu.edu/repos/iuc/anndata_manipulate/anndata_manipulate/0.10.9+galaxy0",
"errors": null,
"id": 25,
"input_connections": {
@@ -894,18 +1040,18 @@
],
"position": {
"left": 559.9999540284473,
- "top": 770
+ "top": 835.3983883124581
},
"post_job_actions": {},
- "tool_id": "toolshed.g2.bx.psu.edu/repos/iuc/anndata_manipulate/anndata_manipulate/0.7.5+galaxy1",
+ "tool_id": "toolshed.g2.bx.psu.edu/repos/iuc/anndata_manipulate/anndata_manipulate/0.10.9+galaxy0",
"tool_shed_repository": {
- "changeset_revision": "3d748954434b",
+ "changeset_revision": "c4209ea387d4",
"name": "anndata_manipulate",
"owner": "iuc",
"tool_shed": "toolshed.g2.bx.psu.edu"
},
- "tool_state": "{\"__input_ext\": \"h5ad\", \"chromInfo\": \"/opt/galaxy/tool-data/shared/ucsc/chrom/?.len\", \"input\": {\"__class__\": \"ConnectedValue\"}, \"manipulate\": {\"function\": \"transpose\", \"__current_case__\": 5}, \"__page__\": null, \"__rerun_remap_job_id__\": null}",
- "tool_version": "0.7.5+galaxy1",
+ "tool_state": "{\"input\": {\"__class__\": \"ConnectedValue\"}, \"manipulate\": {\"function\": \"transpose\", \"__current_case__\": 7}, \"__page__\": null, \"__rerun_remap_job_id__\": null}",
+ "tool_version": "0.10.9+galaxy0",
"type": "tool",
"uuid": "6dea19a9-3530-44d2-97c3-338062560911",
"when": null,
@@ -913,7 +1059,7 @@
},
"26": {
"annotation": "",
- "content_id": "toolshed.g2.bx.psu.edu/repos/iuc/anndata_manipulate/anndata_manipulate/0.7.5+galaxy1",
+ "content_id": "toolshed.g2.bx.psu.edu/repos/iuc/anndata_manipulate/anndata_manipulate/0.10.9+galaxy0",
"errors": null,
"id": 26,
"input_connections": {
@@ -933,18 +1079,18 @@
],
"position": {
"left": 559.9999540284473,
- "top": 924
+ "top": 989.3983883124581
},
"post_job_actions": {},
- "tool_id": "toolshed.g2.bx.psu.edu/repos/iuc/anndata_manipulate/anndata_manipulate/0.7.5+galaxy1",
+ "tool_id": "toolshed.g2.bx.psu.edu/repos/iuc/anndata_manipulate/anndata_manipulate/0.10.9+galaxy0",
"tool_shed_repository": {
- "changeset_revision": "3d748954434b",
+ "changeset_revision": "c4209ea387d4",
"name": "anndata_manipulate",
"owner": "iuc",
"tool_shed": "toolshed.g2.bx.psu.edu"
},
- "tool_state": "{\"__input_ext\": \"h5ad\", \"chromInfo\": \"/opt/galaxy/tool-data/shared/ucsc/chrom/?.len\", \"input\": {\"__class__\": \"ConnectedValue\"}, \"manipulate\": {\"function\": \"transpose\", \"__current_case__\": 5}, \"__page__\": null, \"__rerun_remap_job_id__\": null}",
- "tool_version": "0.7.5+galaxy1",
+ "tool_state": "{\"input\": {\"__class__\": \"ConnectedValue\"}, \"manipulate\": {\"function\": \"transpose\", \"__current_case__\": 7}, \"__page__\": null, \"__rerun_remap_job_id__\": null}",
+ "tool_version": "0.10.9+galaxy0",
"type": "tool",
"uuid": "7741d5fb-301e-4232-b6ab-29cab4ce1550",
"when": null,
@@ -952,7 +1098,7 @@
},
"27": {
"annotation": "",
- "content_id": "toolshed.g2.bx.psu.edu/repos/iuc/anndata_manipulate/anndata_manipulate/0.7.5+galaxy1",
+ "content_id": "toolshed.g2.bx.psu.edu/repos/iuc/anndata_manipulate/anndata_manipulate/0.10.9+galaxy0",
"errors": null,
"id": 27,
"input_connections": {
@@ -972,18 +1118,18 @@
],
"position": {
"left": 559.9999540284473,
- "top": 1078
+ "top": 1143.398388312458
},
"post_job_actions": {},
- "tool_id": "toolshed.g2.bx.psu.edu/repos/iuc/anndata_manipulate/anndata_manipulate/0.7.5+galaxy1",
+ "tool_id": "toolshed.g2.bx.psu.edu/repos/iuc/anndata_manipulate/anndata_manipulate/0.10.9+galaxy0",
"tool_shed_repository": {
- "changeset_revision": "3d748954434b",
+ "changeset_revision": "c4209ea387d4",
"name": "anndata_manipulate",
"owner": "iuc",
"tool_shed": "toolshed.g2.bx.psu.edu"
},
- "tool_state": "{\"__input_ext\": \"h5ad\", \"chromInfo\": \"/opt/galaxy/tool-data/shared/ucsc/chrom/?.len\", \"input\": {\"__class__\": \"ConnectedValue\"}, \"manipulate\": {\"function\": \"transpose\", \"__current_case__\": 5}, \"__page__\": null, \"__rerun_remap_job_id__\": null}",
- "tool_version": "0.7.5+galaxy1",
+ "tool_state": "{\"input\": {\"__class__\": \"ConnectedValue\"}, \"manipulate\": {\"function\": \"transpose\", \"__current_case__\": 7}, \"__page__\": null, \"__rerun_remap_job_id__\": null}",
+ "tool_version": "0.10.9+galaxy0",
"type": "tool",
"uuid": "f8e93ee5-b396-4fff-8501-0c7b96a04732",
"when": null,
@@ -991,7 +1137,7 @@
},
"28": {
"annotation": "",
- "content_id": "toolshed.g2.bx.psu.edu/repos/iuc/anndata_manipulate/anndata_manipulate/0.7.5+galaxy1",
+ "content_id": "toolshed.g2.bx.psu.edu/repos/iuc/anndata_manipulate/anndata_manipulate/0.10.9+galaxy0",
"errors": null,
"id": 28,
"input_connections": {
@@ -1011,18 +1157,18 @@
],
"position": {
"left": 559.9999540284473,
- "top": 1232
+ "top": 1297.398388312458
},
"post_job_actions": {},
- "tool_id": "toolshed.g2.bx.psu.edu/repos/iuc/anndata_manipulate/anndata_manipulate/0.7.5+galaxy1",
+ "tool_id": "toolshed.g2.bx.psu.edu/repos/iuc/anndata_manipulate/anndata_manipulate/0.10.9+galaxy0",
"tool_shed_repository": {
- "changeset_revision": "3d748954434b",
+ "changeset_revision": "c4209ea387d4",
"name": "anndata_manipulate",
"owner": "iuc",
"tool_shed": "toolshed.g2.bx.psu.edu"
},
- "tool_state": "{\"__input_ext\": \"h5ad\", \"chromInfo\": \"/opt/galaxy/tool-data/shared/ucsc/chrom/?.len\", \"input\": {\"__class__\": \"ConnectedValue\"}, \"manipulate\": {\"function\": \"transpose\", \"__current_case__\": 5}, \"__page__\": null, \"__rerun_remap_job_id__\": null}",
- "tool_version": "0.7.5+galaxy1",
+ "tool_state": "{\"input\": {\"__class__\": \"ConnectedValue\"}, \"manipulate\": {\"function\": \"transpose\", \"__current_case__\": 7}, \"__page__\": null, \"__rerun_remap_job_id__\": null}",
+ "tool_version": "0.10.9+galaxy0",
"type": "tool",
"uuid": "50c0a7c0-7c5f-4b6e-8313-716947a8077a",
"when": null,
@@ -1030,7 +1176,7 @@
},
"29": {
"annotation": "",
- "content_id": "toolshed.g2.bx.psu.edu/repos/iuc/anndata_manipulate/anndata_manipulate/0.7.5+galaxy1",
+ "content_id": "toolshed.g2.bx.psu.edu/repos/iuc/anndata_manipulate/anndata_manipulate/0.10.9+galaxy0",
"errors": null,
"id": 29,
"input_connections": {
@@ -1050,18 +1196,18 @@
],
"position": {
"left": 559.9999540284473,
- "top": 1386
+ "top": 1451.398388312458
},
"post_job_actions": {},
- "tool_id": "toolshed.g2.bx.psu.edu/repos/iuc/anndata_manipulate/anndata_manipulate/0.7.5+galaxy1",
+ "tool_id": "toolshed.g2.bx.psu.edu/repos/iuc/anndata_manipulate/anndata_manipulate/0.10.9+galaxy0",
"tool_shed_repository": {
- "changeset_revision": "3d748954434b",
+ "changeset_revision": "c4209ea387d4",
"name": "anndata_manipulate",
"owner": "iuc",
"tool_shed": "toolshed.g2.bx.psu.edu"
},
- "tool_state": "{\"__input_ext\": \"h5ad\", \"chromInfo\": \"/opt/galaxy/tool-data/shared/ucsc/chrom/?.len\", \"input\": {\"__class__\": \"ConnectedValue\"}, \"manipulate\": {\"function\": \"transpose\", \"__current_case__\": 5}, \"__page__\": null, \"__rerun_remap_job_id__\": null}",
- "tool_version": "0.7.5+galaxy1",
+ "tool_state": "{\"input\": {\"__class__\": \"ConnectedValue\"}, \"manipulate\": {\"function\": \"transpose\", \"__current_case__\": 7}, \"__page__\": null, \"__rerun_remap_job_id__\": null}",
+ "tool_version": "0.10.9+galaxy0",
"type": "tool",
"uuid": "0852a076-be4a-4d40-8456-f602e4d98f41",
"when": null,
@@ -1069,7 +1215,7 @@
},
"30": {
"annotation": "",
- "content_id": "toolshed.g2.bx.psu.edu/repos/iuc/anndata_manipulate/anndata_manipulate/0.7.5+galaxy1",
+ "content_id": "toolshed.g2.bx.psu.edu/repos/iuc/anndata_manipulate/anndata_manipulate/0.10.9+galaxy0",
"errors": null,
"id": 30,
"input_connections": {
@@ -1079,44 +1225,49 @@
},
"manipulate|other_adatas": [
{
- "id": 23,
+ "id": 20,
"output_name": "anndata"
},
{
- "id": 24,
+ "id": 21,
"output_name": "anndata"
},
{
- "id": 21,
+ "id": 22,
"output_name": "anndata"
},
{
- "id": 25,
+ "id": 23,
"output_name": "anndata"
},
{
- "id": 22,
+ "id": 24,
"output_name": "anndata"
},
{
- "id": 27,
+ "id": 25,
"output_name": "anndata"
},
{
- "id": 28,
+ "id": 26,
"output_name": "anndata"
},
{
- "id": 26,
+ "id": 27,
"output_name": "anndata"
},
{
- "id": 20,
+ "id": 28,
"output_name": "anndata"
}
]
},
- "inputs": [],
+ "inputs": [
+ {
+ "description": "runtime parameter for tool Manipulate AnnData",
+ "name": "manipulate"
+ }
+ ],
"label": null,
"name": "Manipulate AnnData",
"outputs": [
@@ -1127,7 +1278,7 @@
],
"position": {
"left": 849.9998716765242,
- "top": 622
+ "top": 687.3983883124581
},
"post_job_actions": {
"RenameDatasetActionanndata": {
@@ -1138,15 +1289,15 @@
"output_name": "anndata"
}
},
- "tool_id": "toolshed.g2.bx.psu.edu/repos/iuc/anndata_manipulate/anndata_manipulate/0.7.5+galaxy1",
+ "tool_id": "toolshed.g2.bx.psu.edu/repos/iuc/anndata_manipulate/anndata_manipulate/0.10.9+galaxy0",
"tool_shed_repository": {
- "changeset_revision": "3d748954434b",
+ "changeset_revision": "c4209ea387d4",
"name": "anndata_manipulate",
"owner": "iuc",
"tool_shed": "toolshed.g2.bx.psu.edu"
},
- "tool_state": "{\"__input_ext\": \"h5ad\", \"chromInfo\": \"/opt/galaxy/tool-data/shared/ucsc/chrom/?.len\", \"input\": {\"__class__\": \"ConnectedValue\"}, \"manipulate\": {\"function\": \"concatenate\", \"__current_case__\": 0, \"other_adatas\": {\"__class__\": \"ConnectedValue\"}, \"join\": \"inner\", \"batch_key\": \"batch\", \"index_unique\": \"-\"}, \"__page__\": null, \"__rerun_remap_job_id__\": null}",
- "tool_version": "0.7.5+galaxy1",
+ "tool_state": "{\"input\": {\"__class__\": \"ConnectedValue\"}, \"manipulate\": {\"function\": \"concatenate\", \"__current_case__\": 0, \"other_adatas\": {\"__class__\": \"ConnectedValue\"}, \"join\": \"inner\", \"batch_key\": \"batch\", \"index_unique\": \"-\"}, \"__page__\": null, \"__rerun_remap_job_id__\": null}",
+ "tool_version": "0.10.9+galaxy0",
"type": "tool",
"uuid": "8f439f4f-7495-43fd-a20a-443fb0bb2efa",
"when": null,
@@ -1154,7 +1305,7 @@
},
"31": {
"annotation": "",
- "content_id": "toolshed.g2.bx.psu.edu/repos/iuc/anndata_inspect/anndata_inspect/0.7.5+galaxy1",
+ "content_id": "toolshed.g2.bx.psu.edu/repos/iuc/anndata_inspect/anndata_inspect/0.10.9+galaxy0",
"errors": null,
"id": 31,
"input_connections": {
@@ -1173,8 +1324,8 @@
}
],
"position": {
- "left": 1129.9999080568946,
- "top": 416
+ "left": 1129.9860682890346,
+ "top": 480.08155353594225
},
"post_job_actions": {
"RenameDatasetActionobs": {
@@ -1185,15 +1336,15 @@
"output_name": "obs"
}
},
- "tool_id": "toolshed.g2.bx.psu.edu/repos/iuc/anndata_inspect/anndata_inspect/0.7.5+galaxy1",
+ "tool_id": "toolshed.g2.bx.psu.edu/repos/iuc/anndata_inspect/anndata_inspect/0.10.9+galaxy0",
"tool_shed_repository": {
- "changeset_revision": "ee98d611afc6",
+ "changeset_revision": "14c576ea6148",
"name": "anndata_inspect",
"owner": "iuc",
"tool_shed": "toolshed.g2.bx.psu.edu"
},
- "tool_state": "{\"__input_ext\": \"h5ad\", \"chromInfo\": \"/opt/galaxy/tool-data/shared/ucsc/chrom/?.len\", \"input\": {\"__class__\": \"ConnectedValue\"}, \"inspect\": {\"info\": \"obs\", \"__current_case__\": 3}, \"__page__\": null, \"__rerun_remap_job_id__\": null}",
- "tool_version": "0.7.5+galaxy1",
+ "tool_state": "{\"input\": {\"__class__\": \"ConnectedValue\"}, \"inspect\": {\"info\": \"obs\", \"__current_case__\": 3}, \"__page__\": null, \"__rerun_remap_job_id__\": null}",
+ "tool_version": "0.10.9+galaxy0",
"type": "tool",
"uuid": "d5d36561-8879-456c-83c8-4f4cb07c387e",
"when": null,
@@ -1201,7 +1352,7 @@
},
"32": {
"annotation": "",
- "content_id": "toolshed.g2.bx.psu.edu/repos/bgruening/text_processing/tp_replace_in_column/1.1.3",
+ "content_id": "toolshed.g2.bx.psu.edu/repos/bgruening/text_processing/tp_replace_in_column/9.3+galaxy1",
"errors": null,
"id": 32,
"input_connections": {
@@ -1210,12 +1361,7 @@
"output_name": "obs"
}
},
- "inputs": [
- {
- "description": "runtime parameter for tool Replace Text",
- "name": "infile"
- }
- ],
+ "inputs": [],
"label": null,
"name": "Replace Text",
"outputs": [
@@ -1226,18 +1372,18 @@
],
"position": {
"left": 1409.999944437265,
- "top": 416
+ "top": 481.39838831245817
},
"post_job_actions": {},
- "tool_id": "toolshed.g2.bx.psu.edu/repos/bgruening/text_processing/tp_replace_in_column/1.1.3",
+ "tool_id": "toolshed.g2.bx.psu.edu/repos/bgruening/text_processing/tp_replace_in_column/9.3+galaxy1",
"tool_shed_repository": {
- "changeset_revision": "ddf54b12c295",
+ "changeset_revision": "86755160afbf",
"name": "text_processing",
"owner": "bgruening",
"tool_shed": "toolshed.g2.bx.psu.edu"
},
- "tool_state": "{\"infile\": {\"__class__\": \"RuntimeValue\"}, \"replacements\": [{\"__index__\": 0, \"column\": \"2\", \"find_pattern\": \"0|1\", \"replace_pattern\": \"patient1\"}, {\"__index__\": 1, \"column\": \"2\", \"find_pattern\": \"2|3|4|5|6\", \"replace_pattern\": \"patient2\"}, {\"__index__\": 2, \"column\": \"2\", \"find_pattern\": \"7|8|9\", \"replace_pattern\": \"patient3\"}, {\"__index__\": 3, \"column\": \"2\", \"find_pattern\": \"batch\", \"replace_pattern\": \"patient\"}], \"__page__\": null, \"__rerun_remap_job_id__\": null}",
- "tool_version": "1.1.3",
+ "tool_state": "{\"infile\": {\"__class__\": \"ConnectedValue\"}, \"replacements\": [{\"__index__\": 0, \"column\": \"2\", \"find_pattern\": \"0|1\", \"replace_pattern\": \"patient1\"}, {\"__index__\": 1, \"column\": \"2\", \"find_pattern\": \"2|3|4|5|6\", \"replace_pattern\": \"patient2\"}, {\"__index__\": 2, \"column\": \"2\", \"find_pattern\": \"7|8|9\", \"replace_pattern\": \"patient3\"}, {\"__index__\": 3, \"column\": \"2\", \"find_pattern\": \"batch\", \"replace_pattern\": \"patient\"}], \"__page__\": null, \"__rerun_remap_job_id__\": null}",
+ "tool_version": "9.3+galaxy1",
"type": "tool",
"uuid": "6b9db8dd-5475-4d0d-8721-86c1a295f0da",
"when": null,
@@ -1245,7 +1391,7 @@
},
"33": {
"annotation": "",
- "content_id": "toolshed.g2.bx.psu.edu/repos/bgruening/text_processing/tp_replace_in_column/1.1.3",
+ "content_id": "toolshed.g2.bx.psu.edu/repos/bgruening/text_processing/tp_replace_in_column/9.3+galaxy1",
"errors": null,
"id": 33,
"input_connections": {
@@ -1254,12 +1400,7 @@
"output_name": "obs"
}
},
- "inputs": [
- {
- "description": "runtime parameter for tool Replace Text",
- "name": "infile"
- }
- ],
+ "inputs": [],
"label": null,
"name": "Replace Text",
"outputs": [
@@ -1270,18 +1411,18 @@
],
"position": {
"left": 1409.999944437265,
- "top": 570
+ "top": 635.3983883124581
},
"post_job_actions": {},
- "tool_id": "toolshed.g2.bx.psu.edu/repos/bgruening/text_processing/tp_replace_in_column/1.1.3",
+ "tool_id": "toolshed.g2.bx.psu.edu/repos/bgruening/text_processing/tp_replace_in_column/9.3+galaxy1",
"tool_shed_repository": {
- "changeset_revision": "ddf54b12c295",
+ "changeset_revision": "86755160afbf",
"name": "text_processing",
"owner": "bgruening",
"tool_shed": "toolshed.g2.bx.psu.edu"
},
- "tool_state": "{\"infile\": {\"__class__\": \"RuntimeValue\"}, \"replacements\": [{\"__index__\": 0, \"column\": \"2\", \"find_pattern\": \"2|4|7|8\", \"replace_pattern\": \"poolA\"}, {\"__index__\": 1, \"column\": \"2\", \"find_pattern\": \"3|5|9\", \"replace_pattern\": \"poolB\"}, {\"__index__\": 2, \"column\": \"2\", \"find_pattern\": \"6\", \"replace_pattern\": \"poolC\"}, {\"__index__\": 3, \"column\": \"2\", \"find_pattern\": \"0|1\", \"replace_pattern\": \"NA\"}, {\"__index__\": 4, \"column\": \"2\", \"find_pattern\": \"batch\", \"replace_pattern\": \"replicate\"}], \"__page__\": null, \"__rerun_remap_job_id__\": null}",
- "tool_version": "1.1.3",
+ "tool_state": "{\"infile\": {\"__class__\": \"ConnectedValue\"}, \"replacements\": [{\"__index__\": 0, \"column\": \"2\", \"find_pattern\": \"2|4|7|8\", \"replace_pattern\": \"poolA\"}, {\"__index__\": 1, \"column\": \"2\", \"find_pattern\": \"3|5|9\", \"replace_pattern\": \"poolB\"}, {\"__index__\": 2, \"column\": \"2\", \"find_pattern\": \"6\", \"replace_pattern\": \"poolC\"}, {\"__index__\": 3, \"column\": \"2\", \"find_pattern\": \"0|1\", \"replace_pattern\": \"NA\"}, {\"__index__\": 4, \"column\": \"2\", \"find_pattern\": \"batch\", \"replace_pattern\": \"replicate\"}], \"__page__\": null, \"__rerun_remap_job_id__\": null}",
+ "tool_version": "9.3+galaxy1",
"type": "tool",
"uuid": "e39c581b-79da-4851-b174-cf6b893ba2ee",
"when": null,
@@ -1289,7 +1430,7 @@
},
"34": {
"annotation": "",
- "content_id": "toolshed.g2.bx.psu.edu/repos/bgruening/text_processing/tp_replace_in_column/1.1.3",
+ "content_id": "toolshed.g2.bx.psu.edu/repos/bgruening/text_processing/tp_replace_in_column/9.3+galaxy1",
"errors": null,
"id": 34,
"input_connections": {
@@ -1309,18 +1450,18 @@
],
"position": {
"left": 1689.9999808176353,
- "top": 272
+ "top": 337.39838831245817
},
"post_job_actions": {},
- "tool_id": "toolshed.g2.bx.psu.edu/repos/bgruening/text_processing/tp_replace_in_column/1.1.3",
+ "tool_id": "toolshed.g2.bx.psu.edu/repos/bgruening/text_processing/tp_replace_in_column/9.3+galaxy1",
"tool_shed_repository": {
- "changeset_revision": "ddf54b12c295",
+ "changeset_revision": "86755160afbf",
"name": "text_processing",
"owner": "bgruening",
"tool_shed": "toolshed.g2.bx.psu.edu"
},
- "tool_state": "{\"__input_ext\": \"tabular\", \"chromInfo\": \"/opt/galaxy/tool-data/shared/ucsc/chrom/?.len\", \"infile\": {\"__class__\": \"ConnectedValue\"}, \"replacements\": [{\"__index__\": 0, \"column\": \"2\", \"find_pattern\": \"0\", \"replace_pattern\": \"left-mid\"}, {\"__index__\": 1, \"column\": \"2\", \"find_pattern\": \"1|2|3|8|9\", \"replace_pattern\": \"right-mid\"}, {\"__index__\": 2, \"column\": \"2\", \"find_pattern\": \"4|5|6\", \"replace_pattern\": \"right-apex\"}, {\"__index__\": 3, \"column\": \"2\", \"find_pattern\": \"7\", \"replace_pattern\": \"right-anterior\"}, {\"__index__\": 4, \"column\": \"2\", \"find_pattern\": \"batch\", \"replace_pattern\": \"tumorSpecimen\"}], \"__page__\": null, \"__rerun_remap_job_id__\": null}",
- "tool_version": "1.1.3",
+ "tool_state": "{\"infile\": {\"__class__\": \"ConnectedValue\"}, \"replacements\": [{\"__index__\": 0, \"column\": \"2\", \"find_pattern\": \"0\", \"replace_pattern\": \"left-mid\"}, {\"__index__\": 1, \"column\": \"2\", \"find_pattern\": \"1|2|3|8|9\", \"replace_pattern\": \"right-mid\"}, {\"__index__\": 2, \"column\": \"2\", \"find_pattern\": \"4|5|6\", \"replace_pattern\": \"right-apex\"}, {\"__index__\": 3, \"column\": \"2\", \"find_pattern\": \"7\", \"replace_pattern\": \"right-anterior\"}, {\"__index__\": 4, \"column\": \"2\", \"find_pattern\": \"batch\", \"replace_pattern\": \"tumorSpecimen\"}], \"__page__\": null, \"__rerun_remap_job_id__\": null}",
+ "tool_version": "9.3+galaxy1",
"type": "tool",
"uuid": "0efbddb8-8f61-40b6-878e-1080f313ec5e",
"when": null,
@@ -1328,7 +1469,7 @@
},
"35": {
"annotation": "",
- "content_id": "toolshed.g2.bx.psu.edu/repos/bgruening/text_processing/tp_replace_in_column/1.1.3",
+ "content_id": "toolshed.g2.bx.psu.edu/repos/bgruening/text_processing/tp_replace_in_column/9.3+galaxy1",
"errors": null,
"id": 35,
"input_connections": {
@@ -1347,19 +1488,19 @@
}
],
"position": {
- "left": 1970.0000171980057,
- "top": 194
+ "left": 1968.6831663579067,
+ "top": 259.39836647901654
},
"post_job_actions": {},
- "tool_id": "toolshed.g2.bx.psu.edu/repos/bgruening/text_processing/tp_replace_in_column/1.1.3",
+ "tool_id": "toolshed.g2.bx.psu.edu/repos/bgruening/text_processing/tp_replace_in_column/9.3+galaxy1",
"tool_shed_repository": {
- "changeset_revision": "ddf54b12c295",
+ "changeset_revision": "86755160afbf",
"name": "text_processing",
"owner": "bgruening",
"tool_shed": "toolshed.g2.bx.psu.edu"
},
- "tool_state": "{\"__input_ext\": \"tabular\", \"chromInfo\": \"/opt/galaxy/tool-data/shared/ucsc/chrom/?.len\", \"infile\": {\"__class__\": \"ConnectedValue\"}, \"replacements\": [{\"__index__\": 0, \"column\": \"2\", \"find_pattern\": \"0\", \"replace_pattern\": \"AUG_PB1A\"}, {\"__index__\": 1, \"column\": \"2\", \"find_pattern\": \"1$\", \"replace_pattern\": \"AUG_PB1B\"}, {\"__index__\": 2, \"column\": \"2\", \"find_pattern\": \"(2$)|3\", \"replace_pattern\": \"MAY_PB1A\"}, {\"__index__\": 3, \"column\": \"2\", \"find_pattern\": \"4|5|6\", \"replace_pattern\": \"MAY_PB1B\"}, {\"__index__\": 4, \"column\": \"2\", \"find_pattern\": \"7\", \"replace_pattern\": \"MAY_PB2A\"}, {\"__index__\": 5, \"column\": \"2\", \"find_pattern\": \"8|9\", \"replace_pattern\": \"MAY_PB2B\"}, {\"__index__\": 6, \"column\": \"2\", \"find_pattern\": \"batch\", \"replace_pattern\": \"SpecimenID\"}], \"__page__\": null, \"__rerun_remap_job_id__\": null}",
- "tool_version": "1.1.3",
+ "tool_state": "{\"infile\": {\"__class__\": \"ConnectedValue\"}, \"replacements\": [{\"__index__\": 0, \"column\": \"2\", \"find_pattern\": \"0\", \"replace_pattern\": \"AUG_PB1A\"}, {\"__index__\": 1, \"column\": \"2\", \"find_pattern\": \"1$\", \"replace_pattern\": \"AUG_PB1B\"}, {\"__index__\": 2, \"column\": \"2\", \"find_pattern\": \"(2$)|3\", \"replace_pattern\": \"MAY_PB1A\"}, {\"__index__\": 3, \"column\": \"2\", \"find_pattern\": \"4|5|6\", \"replace_pattern\": \"MAY_PB1B\"}, {\"__index__\": 4, \"column\": \"2\", \"find_pattern\": \"7\", \"replace_pattern\": \"MAY_PB2A\"}, {\"__index__\": 5, \"column\": \"2\", \"find_pattern\": \"8|9\", \"replace_pattern\": \"MAY_PB2B\"}, {\"__index__\": 6, \"column\": \"2\", \"find_pattern\": \"batch\", \"replace_pattern\": \"SpecimenID\"}], \"__page__\": null, \"__rerun_remap_job_id__\": null}",
+ "tool_version": "9.3+galaxy1",
"type": "tool",
"uuid": "03794fc2-ffd5-4315-98c3-f7d225465a78",
"when": null,
@@ -1387,7 +1528,7 @@
],
"position": {
"left": 1690.000040183782,
- "top": 426
+ "top": 491.39838831245817
},
"post_job_actions": {
"RenameDatasetActionout_file1": {
@@ -1428,7 +1569,7 @@
],
"position": {
"left": 1689.9999808176353,
- "top": 580
+ "top": 645.3983883124581
},
"post_job_actions": {
"RenameDatasetActionout_file1": {
@@ -1469,7 +1610,7 @@
],
"position": {
"left": 1970.0000171980057,
- "top": 348
+ "top": 413.39838831245817
},
"post_job_actions": {
"RenameDatasetActionout_file1": {
@@ -1510,7 +1651,7 @@
],
"position": {
"left": 2250.000053578376,
- "top": 216
+ "top": 281.3983883124581
},
"post_job_actions": {
"RenameDatasetActionout_file1": {
@@ -1555,7 +1696,7 @@
],
"position": {
"left": 1970.0000171980057,
- "top": 482
+ "top": 547.3983883124581
},
"post_job_actions": {},
"tool_id": "Paste1",
@@ -1592,7 +1733,7 @@
],
"position": {
"left": 2250.000053578376,
- "top": 350
+ "top": 415.39838831245817
},
"post_job_actions": {},
"tool_id": "Paste1",
@@ -1629,7 +1770,7 @@
],
"position": {
"left": 2530.0000899587462,
- "top": 323
+ "top": 388.39838831245817
},
"post_job_actions": {
"RenameDatasetActionout_file1": {
@@ -1650,7 +1791,7 @@
},
"43": {
"annotation": "",
- "content_id": "toolshed.g2.bx.psu.edu/repos/iuc/anndata_manipulate/anndata_manipulate/0.7.5+galaxy1",
+ "content_id": "toolshed.g2.bx.psu.edu/repos/iuc/anndata_manipulate/anndata_manipulate/0.10.9+galaxy0",
"errors": null,
"id": 43,
"input_connections": {
@@ -1663,7 +1804,12 @@
"output_name": "out_file1"
}
},
- "inputs": [],
+ "inputs": [
+ {
+ "description": "runtime parameter for tool Manipulate AnnData",
+ "name": "manipulate"
+ }
+ ],
"label": null,
"name": "Manipulate AnnData",
"outputs": [
@@ -1674,18 +1820,18 @@
],
"position": {
"left": 2810.0001263391164,
- "top": 605
+ "top": 670.3983883124581
},
"post_job_actions": {},
- "tool_id": "toolshed.g2.bx.psu.edu/repos/iuc/anndata_manipulate/anndata_manipulate/0.7.5+galaxy1",
+ "tool_id": "toolshed.g2.bx.psu.edu/repos/iuc/anndata_manipulate/anndata_manipulate/0.10.9+galaxy0",
"tool_shed_repository": {
- "changeset_revision": "3d748954434b",
+ "changeset_revision": "c4209ea387d4",
"name": "anndata_manipulate",
"owner": "iuc",
"tool_shed": "toolshed.g2.bx.psu.edu"
},
- "tool_state": "{\"__input_ext\": \"h5ad\", \"chromInfo\": \"/opt/galaxy/tool-data/shared/ucsc/chrom/?.len\", \"input\": {\"__class__\": \"ConnectedValue\"}, \"manipulate\": {\"function\": \"add_annotation\", \"__current_case__\": 6, \"var_obs\": \"obs\", \"new_annot\": {\"__class__\": \"ConnectedValue\"}}, \"__page__\": null, \"__rerun_remap_job_id__\": null}",
- "tool_version": "0.7.5+galaxy1",
+ "tool_state": "{\"input\": {\"__class__\": \"ConnectedValue\"}, \"manipulate\": {\"function\": \"add_annotation\", \"__current_case__\": 8, \"var_obs\": \"obs\", \"new_annot\": {\"__class__\": \"ConnectedValue\"}}, \"__page__\": null, \"__rerun_remap_job_id__\": null}",
+ "tool_version": "0.10.9+galaxy0",
"type": "tool",
"uuid": "bafffb92-883e-4954-bec0-55551545a625",
"when": null,
@@ -1693,7 +1839,7 @@
},
"44": {
"annotation": "",
- "content_id": "toolshed.g2.bx.psu.edu/repos/ebi-gxa/scanpy_filter_cells/scanpy_filter_cells/1.8.1+galaxy9",
+ "content_id": "toolshed.g2.bx.psu.edu/repos/ebi-gxa/scanpy_filter_cells/scanpy_filter_cells/1.9.3+galaxy0",
"errors": null,
"id": 44,
"input_connections": {
@@ -1712,19 +1858,19 @@
}
],
"position": {
- "left": 3089.9999252549,
- "top": 610
+ "left": 3090.0002007953785,
+ "top": 673.9778304207833
},
"post_job_actions": {},
- "tool_id": "toolshed.g2.bx.psu.edu/repos/ebi-gxa/scanpy_filter_cells/scanpy_filter_cells/1.8.1+galaxy9",
+ "tool_id": "toolshed.g2.bx.psu.edu/repos/ebi-gxa/scanpy_filter_cells/scanpy_filter_cells/1.9.3+galaxy0",
"tool_shed_repository": {
- "changeset_revision": "e9283529cba1",
+ "changeset_revision": "bf1e0bdec1db",
"name": "scanpy_filter_cells",
"owner": "ebi-gxa",
"tool_shed": "toolshed.g2.bx.psu.edu"
},
- "tool_state": "{\"__input_ext\": \"input\", \"categories\": [], \"chromInfo\": \"/opt/galaxy/tool-data/shared/ucsc/chrom/?.len\", \"export_mtx\": false, \"force_recalc\": false, \"gene_name\": \"_index\", \"input_format\": \"anndata\", \"input_obj_file\": {\"__class__\": \"ConnectedValue\"}, \"output_format\": \"anndata_h5ad\", \"parameters\": [{\"__index__\": 0, \"name\": \"n_genes\", \"min\": \"0.0\", \"max\": \"1000000000.0\"}], \"save_layer\": null, \"save_raw\": false, \"subsets\": [], \"__page__\": null, \"__rerun_remap_job_id__\": null}",
- "tool_version": "1.8.1+galaxy9",
+ "tool_state": "{\"categories\": [], \"export_mtx\": false, \"force_recalc\": false, \"gene_name\": \"_index\", \"input_format\": \"anndata\", \"input_obj_file\": {\"__class__\": \"ConnectedValue\"}, \"output_format\": \"anndata_h5ad\", \"parameters\": [{\"__index__\": 0, \"name\": \"n_genes\", \"min\": \"0.0\", \"max\": \"1000000000.0\"}], \"save_layer\": null, \"save_raw\": false, \"subsets\": [], \"__page__\": null, \"__rerun_remap_job_id__\": null}",
+ "tool_version": "1.9.3+galaxy0",
"type": "tool",
"uuid": "2ae0ca16-f58b-4a3a-bdef-bf66a74b73c2",
"when": null,
@@ -1732,7 +1878,7 @@
},
"45": {
"annotation": "",
- "content_id": "toolshed.g2.bx.psu.edu/repos/ebi-gxa/anndata_ops/anndata_ops/1.8.1+galaxy0",
+ "content_id": "toolshed.g2.bx.psu.edu/repos/ebi-gxa/anndata_ops/anndata_ops/1.9.3+galaxy0",
"errors": null,
"id": 45,
"input_connections": {
@@ -1751,19 +1897,19 @@
}
],
"position": {
- "left": 3369.999842902977,
- "top": 610
+ "left": 3369.985953746747,
+ "top": 674.0815558059719
},
"post_job_actions": {},
- "tool_id": "toolshed.g2.bx.psu.edu/repos/ebi-gxa/anndata_ops/anndata_ops/1.8.1+galaxy0",
+ "tool_id": "toolshed.g2.bx.psu.edu/repos/ebi-gxa/anndata_ops/anndata_ops/1.9.3+galaxy0",
"tool_shed_repository": {
- "changeset_revision": "890fb06a2893",
+ "changeset_revision": "a4774b7b2e85",
"name": "anndata_ops",
"owner": "ebi-gxa",
"tool_shed": "toolshed.g2.bx.psu.edu"
},
- "tool_state": "{\"__input_ext\": \"input\", \"chromInfo\": \"/opt/galaxy/tool-data/shared/ucsc/chrom/?.len\", \"copy_adata_to_raw\": false, \"copy_e\": {\"default\": false, \"__current_case__\": 1}, \"copy_l\": {\"default\": false, \"__current_case__\": 1}, \"copy_o\": {\"default\": false, \"__current_case__\": 1}, \"copy_r\": {\"default\": false, \"__current_case__\": 1}, \"copy_u\": {\"default\": false, \"__current_case__\": 1}, \"copy_x\": {\"default\": false, \"__current_case__\": 1}, \"gene_flags\": [{\"__index__\": 0, \"startswith\": \"MT-\", \"flag\": \"mito\"}], \"gene_symbols_field\": \"index\", \"input_obj_file\": {\"__class__\": \"ConnectedValue\"}, \"modifications\": [], \"output_format\": \"anndata_h5ad\", \"sanitize_varm\": false, \"top_genes\": \"50\", \"__page__\": null, \"__rerun_remap_job_id__\": null}",
- "tool_version": "1.8.1+galaxy0",
+ "tool_state": "{\"add_cell_metadata\": {\"default\": false, \"__current_case__\": 1}, \"copy_adata_to_raw\": false, \"copy_e\": {\"default\": false, \"__current_case__\": 1}, \"copy_l\": {\"default\": false, \"__current_case__\": 1}, \"copy_o\": {\"default\": false, \"__current_case__\": 1}, \"copy_r\": {\"default\": false, \"__current_case__\": 1}, \"copy_u\": {\"default\": false, \"__current_case__\": 1}, \"copy_x\": {\"default\": false, \"__current_case__\": 1}, \"field_unique\": null, \"gene_flags\": [{\"__index__\": 0, \"startswith\": \"MT-\", \"flag\": \"mito\"}], \"gene_symbols_field\": \"index\", \"input_obj_file\": {\"__class__\": \"ConnectedValue\"}, \"modifications\": [], \"output_format\": \"anndata_h5ad\", \"sanitize_varm\": false, \"split_on_obs\": {\"default\": false, \"__current_case__\": 1}, \"swap_layer_to_x\": {\"default\": false, \"__current_case__\": 1}, \"top_genes\": \"50\", \"var_modifications\": [], \"__page__\": null, \"__rerun_remap_job_id__\": null}",
+ "tool_version": "1.9.3+galaxy0",
"type": "tool",
"uuid": "c53d106a-f45b-477d-ab64-ad266c248411",
"when": null,
@@ -1777,7 +1923,7 @@
},
"46": {
"annotation": "",
- "content_id": "toolshed.g2.bx.psu.edu/repos/iuc/anndata_inspect/anndata_inspect/0.7.5+galaxy1",
+ "content_id": "toolshed.g2.bx.psu.edu/repos/iuc/anndata_inspect/anndata_inspect/0.10.9+galaxy0",
"errors": null,
"id": 46,
"input_connections": {
@@ -1797,18 +1943,18 @@
],
"position": {
"left": 3649.9996418187607,
- "top": 642
+ "top": 707.3983883124581
},
"post_job_actions": {},
- "tool_id": "toolshed.g2.bx.psu.edu/repos/iuc/anndata_inspect/anndata_inspect/0.7.5+galaxy1",
+ "tool_id": "toolshed.g2.bx.psu.edu/repos/iuc/anndata_inspect/anndata_inspect/0.10.9+galaxy0",
"tool_shed_repository": {
- "changeset_revision": "ee98d611afc6",
+ "changeset_revision": "14c576ea6148",
"name": "anndata_inspect",
"owner": "iuc",
"tool_shed": "toolshed.g2.bx.psu.edu"
},
- "tool_state": "{\"__input_ext\": \"h5ad\", \"chromInfo\": \"/opt/galaxy/tool-data/shared/ucsc/chrom/?.len\", \"input\": {\"__class__\": \"ConnectedValue\"}, \"inspect\": {\"info\": \"obs\", \"__current_case__\": 3}, \"__page__\": null, \"__rerun_remap_job_id__\": null}",
- "tool_version": "0.7.5+galaxy1",
+ "tool_state": "{\"input\": {\"__class__\": \"ConnectedValue\"}, \"inspect\": {\"info\": \"obs\", \"__current_case__\": 3}, \"__page__\": null, \"__rerun_remap_job_id__\": null}",
+ "tool_version": "0.10.9+galaxy0",
"type": "tool",
"uuid": "60a81f95-32e1-47a7-8a95-07586eb9ed6a",
"when": null,
@@ -1825,6 +1971,6 @@
"name:single-cell",
"name:data-management"
],
- "uuid": "5d73dd95-2534-448a-bb8b-2470bfb1db5a",
- "version": 5
+ "uuid": "da27a6fb-ab9f-4fe7-94b9-1d73e7c0fce7",
+ "version": 4
}
\ No newline at end of file