-
Notifications
You must be signed in to change notification settings - Fork 19.5k
New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
Unpooling and deconvolution #378
Comments
I'm sure this is sufficient Not sure how you want to unpool. Can you elaborate on that? |
|
I need to verify it, but i think theano repeat function can be used for unpooling (upsampling). |
|
I'm currently doing a research involving convolutional autencoder for domain adaptation/transfer learning and using keras thanks to this awesome framework :) What I did is adding the following code in convolutional.py: class Unpooling2D(Layer):
def __init__(self, poolsize=(2, 2), ignore_border=True):
super(Unpooling2D,self).__init__()
self.input = T.tensor4()
self.poolsize = poolsize
self.ignore_border = ignore_border
def get_output(self, train):
X = self.get_input(train)
s1 = self.poolsize[0]
s2 = self.poolsize[1]
output = X.repeat(s1, axis=2).repeat(s2, axis=3)
return output
def get_config(self):
return {"name":self.__class__.__name__,
"poolsize":self.poolsize,
"ignore_border":self.ignore_border}It works well, at least, for my needs. This unpooling strategy is according to this blog post: https://swarbrickjones.wordpress.com/2015/04/29/convolutional-autoencoders-in-pythontheanolasagne/ |
|
thanks @ghif for the unpooling layer. what about deconvolutional layers? has this been implemented in keras? |
|
@Tgaaly Did you ever figure this out? I'm willing to put the effort into making a CAE and post a test example on github if you can help me. |
|
I am trying to create convolutional autoencoder for temporal sequences using Keras. ae = Sequential()
encoder = containers.Sequential([Convolution1D(5, 5, border_mode='valid', input_dim=39, activation='tanh', input_length=39), Flatten(), Dense(5)])
decoder = containers.Sequential([Convolution1D(5, 5, border_mode='valid', input_dim=5, activation='tanh', input_length=5), Flatten(), Dense(39)])
ae.add(AutoEncoder(encoder=encoder, decoder=decoder,
output_reconstruction=False))
ae.compile(loss='mean_squared_error', optimizer=RMSprop())
ae.fit(X_train, X_train, batch_size=32, verbose=1)I am getting the following error at the 3rd line where decoder is being initialized: Please help me resolve the issue |
|
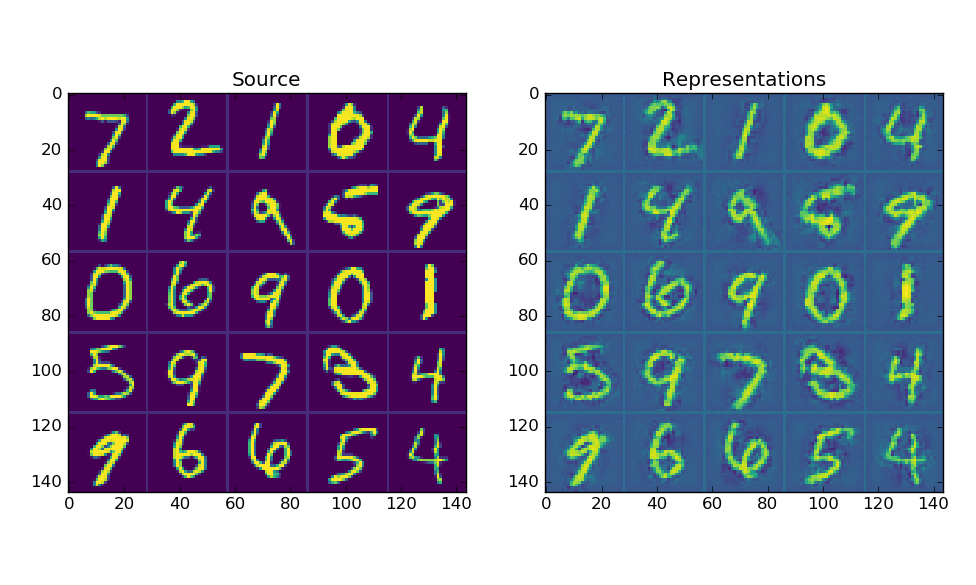
I've prepared the proof of concept of a convolutional autoencoder with weight-sharing between convolutional-deconvolutional layer pairs and maxpool/depool layers with activated neurons sharing. The code is very sketchy at this point, however it illustrates the whole idea. Here's autoencoder's representations of MNIST: I hope it might be useful for someone. |
|
@nanopony awesome demo, thanks for making it. I've been playing with it a bit and noticed something unexpected. If you drop the number of elements in the dense layer to a single neuron you still get reasonable outputs. I was expecting that to have thrown out way too much information to be able to reconstruct the input. Is this a misunderstanding of the architecture on my part? Update: My guess is that you intended the output of grad to be a scalar, but in reality it's redrawing the input to |
|
@kevinthedestroyr That's very odd behavior, since I was getting more blurred and distorted representations when I was reducing size of a bottleneck. Perhaps, if share your code as a gist, I'll check it out on my setup.
The aim of this function is the following. For simplicity sake, assume the maxpool layer gets (2x2) as an input: Lets sum all the outputs ( I am pretty new to Theano as well, so I might have get something wrong, thus detailed |
|
@nanopony I forked to illustrate the behavior I was seeing. Also here's the change I made: As you can see, I replaced the Dense layer with an untrainable single-neuron Dense layer with all weights set to 1. This should act as a wall so that no information should be able to pass through it and you end up with uniform outputs regardless of inputs. But you end up with this unexpected result: Whereas, if you comment out the T.grad line in autoencoder_layers.py you end up with the expected result of uniform output: |
|
@nanopony This is awesome. I'm trying to use the package for arrays of a different size, other than 28X28 pixels. But I'm getting the error msg below. My code is:
|
|
@kevinthedestroyr Thank you for prompt response! I've successfully repeated the behavior. After a bit of tinkering around debug.print, I think I actually have the explanation for observed effect. The single-neuron does works as a wall: provided it sums up 6272 inputs, it is very likely to be active and passing 1 to output in most cases. But, the structural information slips through the connection between MaxPool and DePool in the following way:
Thus, even single-neuron loses all the information, depool still recovers maximums of each 2x2 cells, providing at least something for deconv layer to recover it even further. And therefore, if one disable this connection, information indeed won't pass and we will get the same patterns :) Thank you for this observation! |
|
@talrozen I need to see the actual code of the model to troubleshoot. If you'd like we can discuss it in details at my repo, so this ticket won't be overburdened with tech details. The another issue I see is that I used python3, thus you might be required to change super call to accommodate lack of super shorthand in py2.x |
|
Is it possible to create multi-layers convolutional autoencoder ?? I'm trying to do it but it's not a sucess and I can't find any code with it. |
|
Any update on creating multi-layers convolution autoencoder? |
|
Hi everybody. I'm currently trying to figure out how to build a proper decoder so to preserve the dimensions of the encoding outputs. The problem I found is specifically in the upsampling: in the encoding max-pooling, if the input is not perfectly divisible by the pooling factor, the output will be approximated depending on the 'padding' mode: 'valid' padding: 'same' padding: On the other hand, after the upsampling, the output will be equal to the input multiplied by the upsampling factor (which I keep equal to the pooling size (3,3)), therefore (24,24) for 'same' and (27,27) for 'valid': in both cases they are not the (25,25) I need. After a bit of work on it I found this solution can be used: we can fix the pooling padding to 'same' so that the upsampling will always give a dimension bigger than the one needed and than we could crop it. In my code I collect the pre-pooling dimensions in a list called 'pooling_input_dimension' (for example it could be [(25, 25)]), and then, after each upsampling layer 'x', I calculate the crop dimension this way: Here I needed the 'if' check so to make it work with Cropping1D, since I give the variable 'crop' as input to a 'cropping_layer' which comes already set as Keras Cropping1D or Cropping2D depending on the input dimension. Let me know if any of you found better solutions! |
|
Seems now unpooling with indices can be done using https://github.com/PavlosMelissinos/enet-keras/blob/master/src/models/layers/pooling.py |
For downstream use cases, nice to be able to have this in the ops layer, instead of being forced to reach into backend.
For downstream use cases, nice to be able to have this in the ops layer, instead of being forced to reach into backend.


I want to create convolutional autoencoder and need deconv and unpooling layers.
Is there a way to do that with Keras?
In general i was thinking of using theano repeat function for unpooling
and regular keras conv layer for deconv
What do you think?
The text was updated successfully, but these errors were encountered: