| jupyter | ||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
In the last lesson, we introduced the NIfTI. NIfTI is one of the most ubiquitous file formats for storing neuroimaging data. We'll cover a few details to get started working with them. If you're interested in learning more about NIfTI images, we highly recommend this blog post about the NIfTI format.
NiBabel is a Python package for reading and writing neuroimaging data. To learn more about how NiBabel handles NIfTIs, check out the Working with NIfTI images page of the NiBabel documentation.
import nibabel as nibFirst, use the load() function to create a NiBabel image object from a NIfTI file. We'll load in an example T1w image from the zip file we just downloaded.
t1_img = nib.load("../data/dicom_examples/nii/dcm_anat_ses-01_T1w_20190219111436_5.nii.gz")Loading in a NIfTI file with nibabel gives us a special type of data object which encodes all the information in the file. Each bit of information is called an attribute in Python's terminology. To see all of these attributes, type t1_img. and Tab.
There are three main attributes that we'll discuss today:
1. Header: contains metadata about the image, such as image dimensions, data type, etc.
t1_hdr = t1_img.header
print(t1_hdr)t1_hdr is a Python dictionary. Dictionaries are containers that hold pairs of objects - keys and values. Let's take a look at all of the keys.
Similar to t1_img in which attributes can be accessed by typing t1_img. and hitting Tab, you can do the same with t1_hdr. In particular, we'll be using a method belonging to t1_hdr that will allow you to view the keys associated with it.
t1_hdr.keys()Notice that methods require you to include () at the end of them whereas attributes do not.
The key difference between a method and an attribute is:
- Attributes are stored values kept within an object
- Methods are processes that we can run using the object. Usually a method takes attributes, performs an operation on them, then returns it for you to use.
When you type in t1_img. followed by Tab, you'll see that attributes are highlighted in orange and methods highlighted in blue .
The output above is a list of keys you can use from t1_hdr to access values. We can access the value stored by a given key by typing:
t1_hdr['<key_name>']EXERCISE: Extract the value of pixdim from t1_hdr
t1_hdr['pixdim']In addition to metadata embedded in the NIfTI header, the T1w image also has a corresponding JSON file with additional scan acquisition details. Using the JSON file to store this information is a concept added by BIDS (which we'll cover in the next lesson) to log the important bits of information that traditionally get excluded from the NIfTI header.
Let's take a look at it below:
import json
with open("../data/dicom_examples/nii/dcm_anat_ses-01_T1w_20190219111436_5.json", "r") as f:
t1_metadata = json.load(f)
t1_metadataThe additional metadata are also in the form of a Python dictionary.
EXERCISE: Extract the value of SliceThickness from t1_metadata similar to how you did previously for t1_hdr.
t1_metadata["SliceThickness"]As you've seen above, the header contains useful information that gives us information about the properties (metadata) associated with the MR data we've loaded in. Now we'll move in to loading the actual image data itself. We can achieve this by using the method called t1_img.get_fdata().
t1_data = t1_img.get_fdata()
t1_dataWhat type of data is this exactly? We can determine this by calling the type() function on t1_data.
type(t1_data)The data is a multidimensional array representing the image data. In Python, an array is used to store lists of numerical data into something like a table.
EXERCISE: Let's check out some attributes of the array. How can we see the number of dimensions in the t1_data array? What about the how big each dimension is (shape)? Once again, all of the attributes of the array can be seen by typing t1_data. and Tab.
t1_data.ndimt1_data contains 3 dimensions. You can think of the data as a 3D version of a picture (more accurately, a volume).
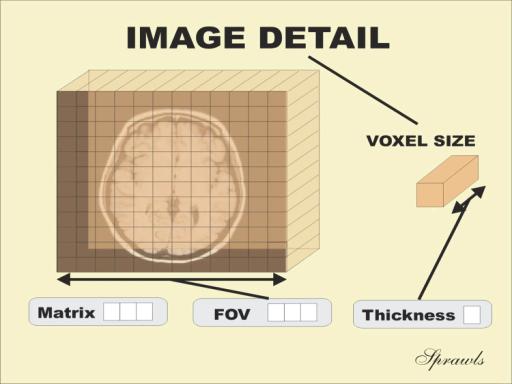
While typical 2D pictures are made out of squares called pixels, a 3D MR image is made up of 3D cubes called voxels.
t1_data.shapeThe 3 numbers given here represent the number of values along a respective dimension (x,y,z). This brain was scanned in 192 slices with a resolution of 256 x 256 voxels per slice. That means there are:
Let's see the type of data inside of the array.
t1_data.dtypeThis tells us that each element in the array (or voxel) is a floating-point number.
The data type of an image controls the range of possible intensities. As the number of possible values increases, the amount of space the image takes up in memory also increases.
import numpy as np
print(np.min(t1_data))
print(np.max(t1_data))For our data, the range of intensity values goes from 0 (black) to more positive digits (whiter).
How do we examine what value a particular voxel is? We can inspect the value of a voxel by selecting an index as follows:
data[x,y,z]So for example we can inspect a voxel at coordinates (10,20,3) by doing the following:
t1_data[9, 19, 2]NOTE: Python uses zero-based indexing. The first item in the array is item 0. The second item is item 1, the third is item 2, etc.
This yields a single value representing the intensity of the signal at a particular voxel! Next we'll see how to not just pull one voxel but a slice or an array of voxels for visualization and analysis!
Slicing does exactly what it seems to imply. Giving our 3D volume, we pull out a 2D slice of our data. Here's an example of slicing from left to right (sagittal slicing):
This gif is a series of 2D images or slices moving from left to right.
Let's pull the 50th slice in the x axis.
x_slice = t1_data[49, :, :]This is similar to the indexing we did before to pull out a single voxel. However, instead of providing a value for each axis, the : indicates that we want to grab all values from that particular axis.
EXERCISE: Now try selecting the 80th slice from the y axis.
y_slice = t1_data[:, 79, :]EXERCISE: Finally try grabbing the 100th slice from the z axis.
z_slice = t1_data[:, :, 99]We've been slicing and dicing brain images but we have no idea what they look like! In the next section we'll show you how you can visualize brain slices!
We previously inspected the signal intensity of the voxel at coordinates (10,20,3). Let's see what out data looks like when we slice it at this location. We've already indexed the data at each x, y, and z axis. Let's use matplotlib, Python's standard plotting library.
import matplotlib.pyplot as plt
%matplotlib inline
slices = [x_slice, y_slice, z_slice]
fig, axes = plt.subplots(1, len(slices))
for i, slice in enumerate(slices):
axes[i].imshow(slice.T, cmap="gray", origin="lower")from nilearn import plotting
plotting.view_img(t1_img, black_bg=True, colorbar=False, cmap="gray", symmetric_cmap=False, vmin=0)An upgrade from matplotlib is nilearn, which provides several advanced plotting features for neuroimaging data.
from nilearn import plotting
plotting.view_img(t1_img, black_bg=True, colorbar=False, cmap="gray", symmetric_cmap=False, vmin=0)Now, we're going to step away from discussing our data and talk about the final important attribute of a NIfTI.
3. Affine: tells the position of the image array data in a reference space
The final important piece of metadata associated with an image file is the affine matrix. Below is the affine matrix for our data.
t1_affine = t1_img.affine
t1_affineTo explain this concept, recall that we referred to coordinates in our data as (x,y,z) coordinates such that:
- x is the first dimension of
t1_data - y is the second dimension of
t1_data - z is the third dimension of
t1_data
Although this tells us how to access our data in terms of voxels in a 3D volume, it doesn't tell us much about the actual dimensions in our data (centimetres, right or left, up or down, back or front). The affine matrix allows us to translate between voxel coordinates (x,y,z) and world space coordinates in (left/right,bottom/top,back/front). An important thing to note is that in reality in which order you have:
- left/right
- bottom/top
- back/front
Depends on how you've constructed the affine matrix, but for the data we're dealing with it always refers to:
- Right
- Anterior
- Superior
Applying the affine matrix (t1_affine) is done through using a linear map (matrix multiplication) on voxel coordinates (defined in t1_data).
The concept of an affine matrix may seem confusing at first but an example might help gain an intuition:
Suppose we have two voxels located at the the following coordinates:
And we wanted to know what the distances between these two voxels are in terms of real world distances (millimetres). This information cannot be derived from using voxel coordinates so we turn to the affine matrix.
Now, the affine matrix we'll be using happens to be encoded in RAS. That means once we apply the matrix our coordinates are as follows:
So increasing a coordinate value in the first dimension corresponds to moving to the right of the person being scanned.
Applying our affine matrix yields the following coordinates:
This means that:
- Voxel 1 is
$90.23-10.25= 79.98$ in the R axis. Positive values mean move right - Voxel 1 is
$0.2-30.5= -30.3$ in the A axis. Negative values mean move posterior - Voxel 1 is
$2.15-9.2= -7.05$ in the S axis. Negatve values mean move inferior
This covers the basics of how NIfTI data and metadata are stored and organized in the context of Python. In the next segment we'll talk a bit about an increasingly important component of MR data analysis - data organization. This is a key component to reproducible analysis and so we'll spend a bit of time here.