Specify step and flyscan paths in a serializable, efficient and Pythonic way using combinations of:
- Specs like Line or Spiral
- Optionally Snaking
- Zip, Product and Concat to compose
- Masks with multiple Regions to restrict
Serialize the Spec rather than the expanded Path and reconstruct it on the server. It can them be iterated over like a cycler, or a stack of scan Frames can be produced and expanded Paths created to consume chunk by chunk.
| Source | https://github.com/bluesky/scanspec |
|---|---|
| PyPI | pip install scanspec |
| Docker | docker run ghcr.io/bluesky/scanspec:latest |
| Documentation | https://bluesky.github.io/scanspec |
| Releases | https://github.com/bluesky/scanspec/releases |
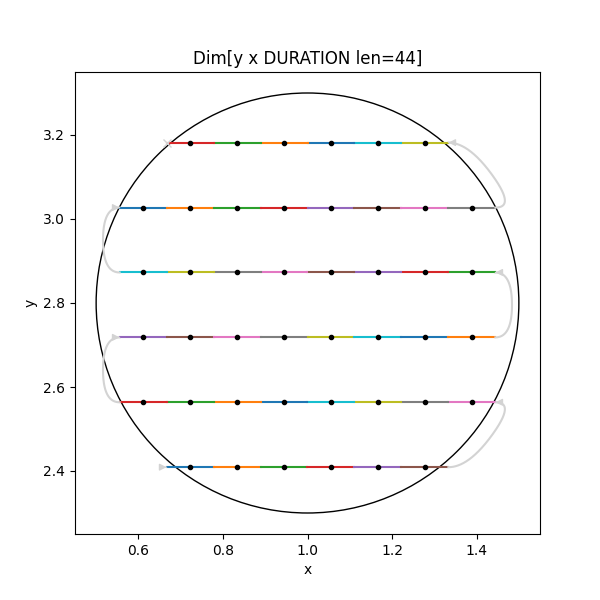
An example ScanSpec of a 2D snaked grid flyscan inside a circle spending 0.4s at each point:
from scanspec.specs import Line, fly
from scanspec.regions import Circle
grid = Line(y, 2.1, 3.8, 12) * ~Line(x, 0.5, 1.5, 10)
spec = fly(grid, 0.4) & Circle(x, y, 1.0, 2.8, radius=0.5)Which when plotted looks like:
Scan points can be iterated through directly for convenience:
for point in spec.midpoints():
print(point)
# ...
# {'y': 3.1818181818181817, 'x': 0.8333333333333333, 'DURATION': 0.4}
# {'y': 3.1818181818181817, 'x': 0.7222222222222222, 'DURATION': 0.4}or a Path created from the stack of Frames and chunks of a given length consumed from it for performance:
from scanspec.core import Path
stack = spec.calculate()
len(stack[0]) # 44
stack[0].axes() # ['y', 'x', 'DURATION']
path = Path(stack, start=5, num=30)
chunk = path.consume(10)
chunk.midpoints # {'x': <ndarray len=10>, 'y': <ndarray len=10>, 'DURATION': <ndarray len=10>}
chunk.upper # bounds are same dimensionality as positionsSee https://bluesky.github.io/scanspec for more detailed documentation.