-
Notifications
You must be signed in to change notification settings - Fork 2
Start using PeptideShaker Online
Before you start using PeptideShaker Online, you will have to get a Galaxy user API key. Although PeptideShaker Online allows you to execute tasks anonymously, your data will then be shared with anybody using the application. So, apart from testing purposes, before running tasks it is highly recommended to use a specific user in order to have your own data history and be able to manage it and execute tasks associated only to your user.
Once you have registered your user using the register user form of your Galaxy GUI instance (please note that this user should not be the same as the user configured for managing the Galaxy instance, and in this case should not have admin permissions), you can generate an API key for it. API keys allow the use of your Galaxy user account programmatically without sharing your private password. This can be easily done when logged in with your user, accessing your user preferences, and clicking "Manage API Key" - "Create a new Key".
Please keep this key in a safe place and then open the PeptideShaker Online website with your browser. Once the main PeptideShaker Online page is shown, click the bottom-right button called "Galaxy Login" and enter the previously generated API Key. Once you click on "Login" button you will be returned to the main PeptideShaker Online page logged in with your Galaxy user ready to use all of its features.
When a user has successfully log in with their private API key, they can upload their own FASTA files and spectrum files through the Search tab. Currently, the supported spectrum input formats are mgf and mzML for identification and Thermo raw files for both identification and quantification.
Users can also upload and visualize their own processed data files using the Upload Your Own Project Files tab located under the Visualize Data tab. Users need to upload FASTA, Proteins, and Peptides files. Please use Sample Files as a template.
The users select the dataset from the "Projects Overview" tab then "Dataset Overview" will be automatically displayed, the tab contest of the following:
- A. Protein inference and protein validation filters
- B. Chromosome filter (chromosome mapping extracted from Uniprot)
- C. Post-translational modifications filter
- D. Number of peptides filter
- E. Number of peptide-to-spectrum filter
- F. Protein coverage filter
- G. Protein intensity filter
- H. Protein table with the currently filtered results.
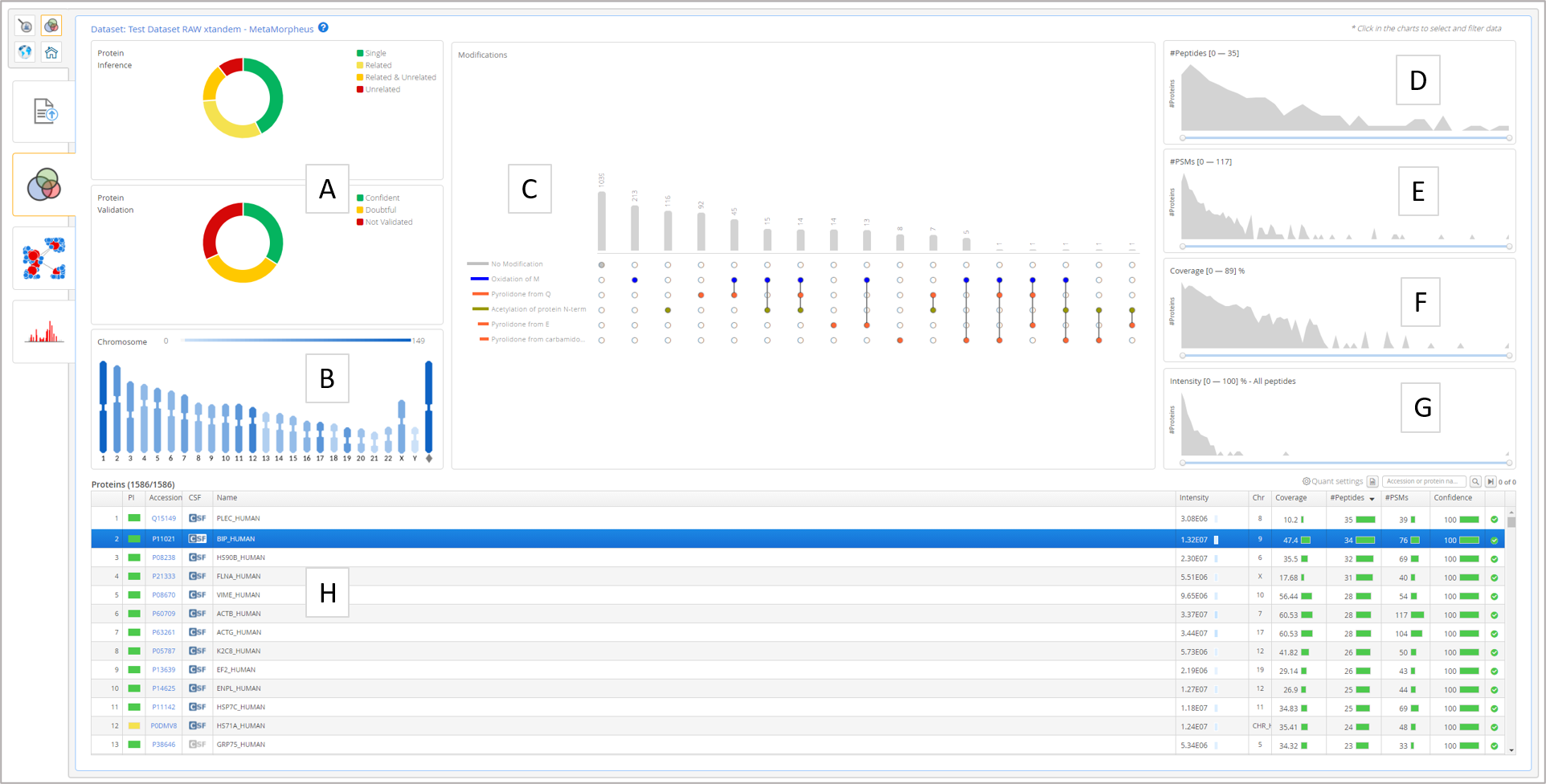
The user filters the dataset at the Dataset Overview level to find and select a protein (group) for closer inspection at the Protein Overview level. Upon selecting the protein (group), the "Protein Visualization" tab will be activated and the user can navigate to it.

The "Protein Visualization" tab includes the following:
- A. Protein-peptide (or proteoform based on information from Reactom database)
- B. Protein 3D structure (using LiteMol 3D visualization)
- C. Protein coverage table that shows the distribution of the peptides and their location.
Here the user selects a peptide from the protein-peptide network or protein coverage table in order to see the peptide and spectrum details at the Peptide-Spectrum Matches level. Upon selecting the interesting peptide, the "Peptide-to-Spectrum Matches" tab will be activated and the user can navigate to it.
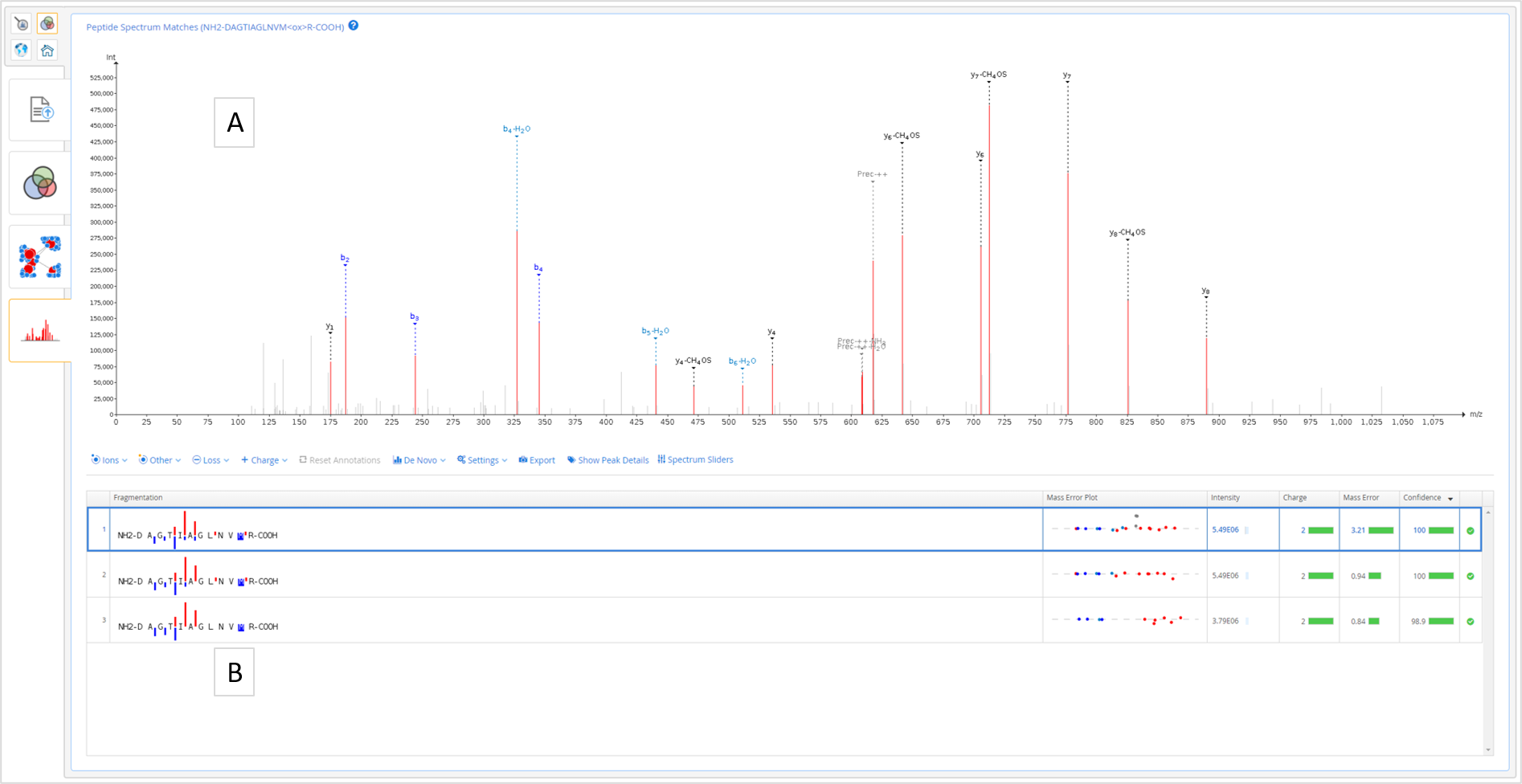
The "Peptide-to-Spectrum Matches" tab consists of the following:
- A. Interactive spectrum viewer where the user can zoom in or out and use all the features including _denovo _ sequencing
- B. Peptide-to-spectrum matches table.