diff --git a/README.md b/README.md
index b794087..3e7952d 100644
--- a/README.md
+++ b/README.md
@@ -5,7 +5,13 @@
`ConformalPrediction.jl` is a package for Uncertainty Quantification (UQ) through Conformal Prediction (CP) in Julia. It is designed to work with supervised models trained in [MLJ](https://alan-turing-institute.github.io/MLJ.jl/dev/) (Blaom et al. 2020). Conformal Prediction is distribution-free, easy-to-understand, easy-to-use and model-agnostic.
-# 📖 Background
+## 🏃 Quick Tour
+
+> First time here? Take a quick interactive [tour](https://binder.plutojl.org/v0.19.12/open?url=https%253A%252F%252Fraw.githubusercontent.com%252Fpat-alt%252FConformalPrediction.jl%252Fmain%252Fdev%252Fquick_tour%252Fnotebook.jl) to see what this package can do.
+
+The [link](https://binder.plutojl.org/v0.19.12/open?url=https%253A%252F%252Fraw.githubusercontent.com%252Fpat-alt%252FConformalPrediction.jl%252Fmain%252Fdev%252Fquick_tour%252Fnotebook.jl) takes you to a `Pluto.jl` notebook hosted on binder.org. In my own experience, this may take a while to load, certainly enough time to get yourself a hot beverage ☕. Alternatively, you can clone this repo and run the notebook contained in `dev/quick-tour` locally or skip the tour for now and read on here.
+
+## 📖 Background
Conformal Prediction is a scalable frequentist approach to uncertainty quantification and coverage control. It promises to be an easy-to-understand, distribution-free and model-agnostic way to generate statistically rigorous uncertainty estimates. Interestingly, it can even be used to complement Bayesian methods.
@@ -82,13 +88,13 @@ ŷ[1:show_first]
```
5-element Vector{Tuple{Float64, Float64}}:
- (0.325476135568554, 2.5420611849529986)
- (-0.8093221495344456, 1.513229072277355)
- (0.24246467414510378, 2.531848511672005)
- (-0.37629465789570005, 1.9144457517084361)
- (-0.5411423339519135, 1.712803571302072)
+ (0.5113539995719073, 2.7791173590180245)
+ (0.15501260477711076, 2.491986075800726)
+ (-0.32783606947941524, 1.9302674946467009)
+ (-0.13732511816023366, 2.141708832043786)
+ (0.5089900787456267, 2.7771571126470387)
-For simple models like this one, we can call `Plots.plot` on our instance, fit result and data to generate the chart below:
+For simple models like this one, we can call a custom `Plots` recipe on our instance, fit result and data to generate the chart below:
``` julia
using Plots
@@ -117,13 +123,13 @@ println("SSC: $(round(_eval.measurement[2], digits=3))")
┌───────────────────────────────────────────────────────────┬───────────┬───────
│ measure │ operation │ meas ⋯
├───────────────────────────────────────────────────────────┼───────────┼───────
- │ emp_coverage (generic function with 1 method) │ predict │ 0.95 ⋯
- │ size_stratified_coverage (generic function with 1 method) │ predict │ 0.90 ⋯
+ │ emp_coverage (generic function with 1 method) │ predict │ 0.94 ⋯
+ │ size_stratified_coverage (generic function with 1 method) │ predict │ 0.76 ⋯
└───────────────────────────────────────────────────────────┴───────────┴───────
3 columns omitted
- Empirical coverage: 0.957
- SSC: 0.907
+ Empirical coverage: 0.947
+ SSC: 0.762
## 🔁 Status
diff --git a/README_files/figure-commonmark/cell-11-output-1.svg b/README_files/figure-commonmark/cell-11-output-1.svg
index 7a0fe1b..cef734e 100644
--- a/README_files/figure-commonmark/cell-11-output-1.svg
+++ b/README_files/figure-commonmark/cell-11-output-1.svg
@@ -1,119 +1,125 @@
diff --git a/README_files/figure-commonmark/cell-7-output-1.svg b/README_files/figure-commonmark/cell-7-output-1.svg
index 716055e..3c6b8b8 100644
--- a/README_files/figure-commonmark/cell-7-output-1.svg
+++ b/README_files/figure-commonmark/cell-7-output-1.svg
@@ -1,439 +1,496 @@
+
diff --git a/_freeze/docs/src/index/execute-results/md.json b/_freeze/docs/src/index/execute-results/md.json
index 834cf61..2ca202c 100644
--- a/_freeze/docs/src/index/execute-results/md.json
+++ b/_freeze/docs/src/index/execute-results/md.json
@@ -1,7 +1,7 @@
{
"hash": "34358001c3f8363312cf01a952052809",
"result": {
- "markdown": "```@meta\nCurrentModule = ConformalPrediction\n```\n\n# ConformalPrediction\n\nDocumentation for [ConformalPrediction.jl](https://github.com/pat-alt/ConformalPrediction.jl).\n\n\n\n`ConformalPrediction.jl` is a package for Uncertainty Quantification (UQ) through Conformal Prediction (CP) in Julia. It is designed to work with supervised models trained in [MLJ](https://alan-turing-institute.github.io/MLJ.jl/dev/) [@blaom2020mlj]. Conformal Prediction is distribution-free, easy-to-understand, easy-to-use and model-agnostic. \n\n# 📖 Background\n\nConformal Prediction is a scalable frequentist approach to uncertainty quantification and coverage control. It promises to be an easy-to-understand, distribution-free and model-agnostic way to generate statistically rigorous uncertainty estimates. Interestingly, it can even be used to complement Bayesian methods.\n\nThe animation below is lifted from a small blog post that introduces the topic and the package ([[TDS](https://towardsdatascience.com/conformal-prediction-in-julia-351b81309e30)], [[Quarto](https://www.paltmeyer.com/blog/posts/conformal-prediction/#fig-anim)]). It shows conformal prediction sets for two different samples and changing coverage rates. Standard conformal classifiers produce set-valued predictions: for ambiguous samples these sets are typically large (for high coverage) or empty (for low coverage).\n\n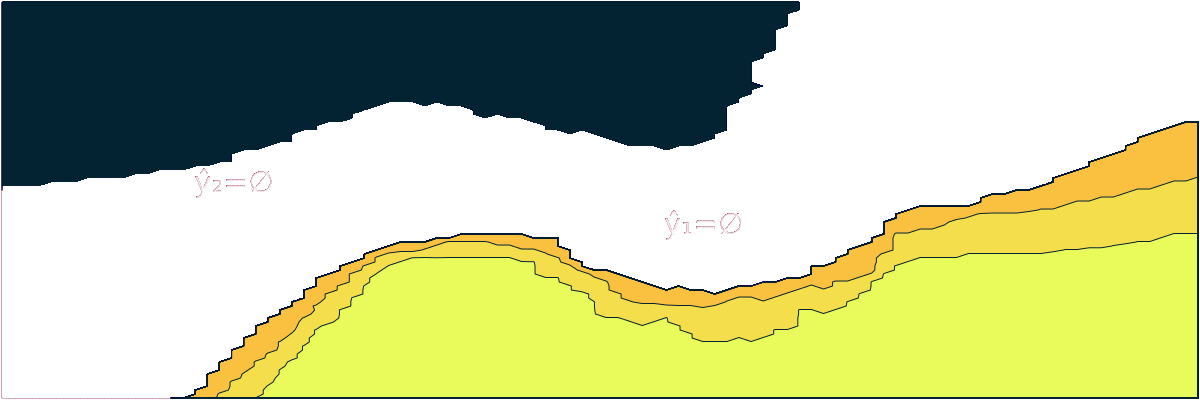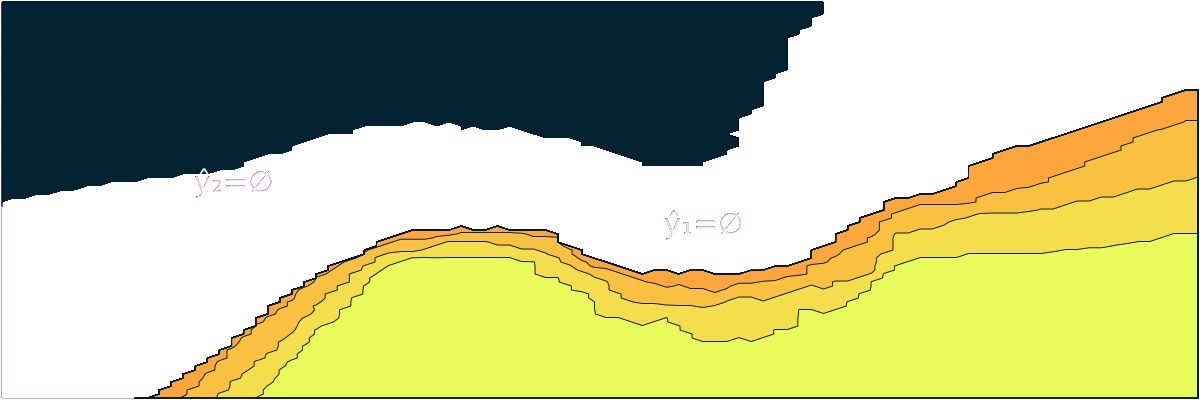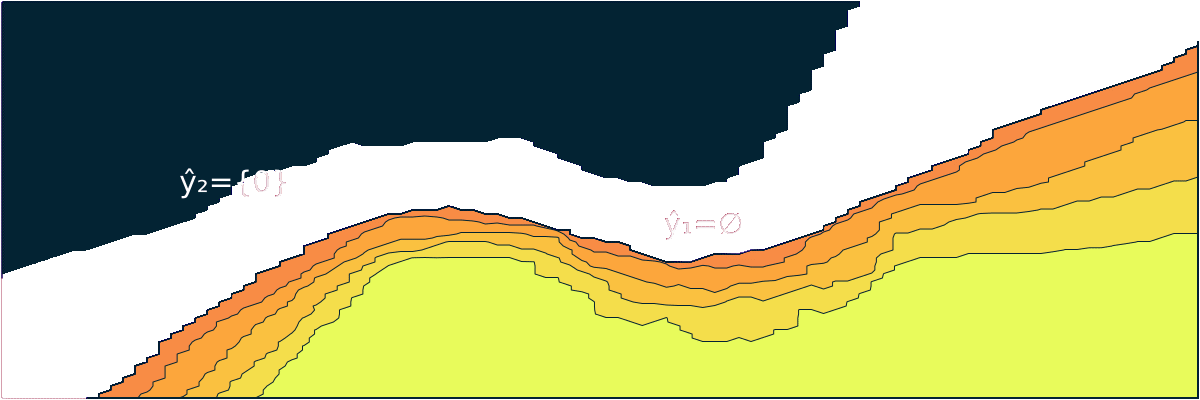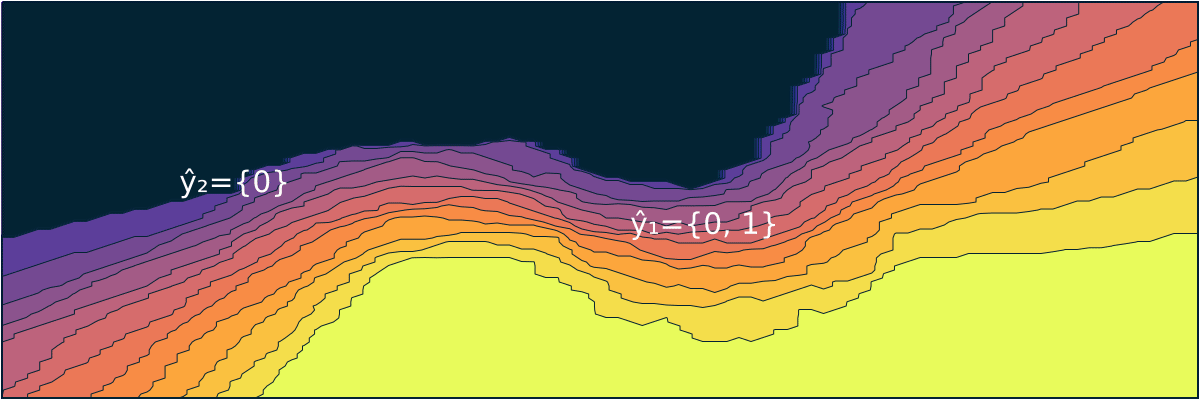\n\n## 🚩 Installation \n\nYou can install the latest stable release from the general registry:\n\n```{.julia}\nusing Pkg\nPkg.add(\"ConformalPrediction\")\n```\n\nThe development version can be installed as follows:\n\n```{.julia}\nusing Pkg\nPkg.add(url=\"https://github.com/pat-alt/ConformalPrediction.jl\")\n```\n\n## 🔍 Usage Example \n\nTo illustrate the intended use of the package, let's have a quick look at a simple regression problem. We first generate some synthetic data and then determine indices for our training and test data using [MLJ](https://alan-turing-institute.github.io/MLJ.jl/dev/):\n\n::: {.cell execution_count=2}\n``` {.julia .cell-code}\nusing MLJ\n\n# Inputs:\nN = 600\nxmax = 3.0\nusing Distributions\nd = Uniform(-xmax, xmax)\nX = rand(d, N)\nX = reshape(X, :, 1)\n\n# Outputs:\nnoise = 0.5\nfun(X) = X * sin(X)\nε = randn(N) .* noise\ny = @.(fun(X)) + ε\ny = vec(y)\n\n# Partition:\ntrain, test = partition(eachindex(y), 0.4, 0.4, shuffle=true)\n```\n:::\n\n\nWe then import a decision-tree based regressor ([`EvoTrees.jl`](https://github.com/Evovest/EvoTrees.jl)) following the standard [MLJ](https://alan-turing-institute.github.io/MLJ.jl/dev/) procedure.\n\n::: {.cell execution_count=3}\n``` {.julia .cell-code}\nEvoTreeRegressor = @load EvoTreeRegressor pkg=EvoTrees\nmodel = EvoTreeRegressor(rounds=100) \n```\n:::\n\n\nTo turn our conventional model into a conformal model, we just need to declare it as such by using `conformal_model` wrapper function. The generated conformal model instance can wrapped in data to create a *machine*. Finally, we proceed by fitting the machine on training data using the generic `fit!` method:\n\n::: {.cell execution_count=4}\n``` {.julia .cell-code}\nusing ConformalPrediction\nconf_model = conformal_model(model; method=:jackknife_plus)\nmach = machine(conf_model, X, y)\nfit!(mach, rows=train)\n```\n:::\n\n\nPredictions can then be computed using the generic `predict` method. The code below produces predictions for the first `n` samples. Each tuple contains the lower and upper bound for the prediction interval. \n\n::: {.cell execution_count=5}\n``` {.julia .cell-code}\nshow_first = 5\nXtest = selectrows(X, test)\nytest = y[test]\nŷ = predict(mach, Xtest)\nŷ[1:show_first]\n```\n\n::: {.cell-output .cell-output-display execution_count=30}\n```\n5-element Vector{Tuple{Float64, Float64}}:\n (0.5113539995719073, 2.7791173590180245)\n (0.15501260477711076, 2.491986075800726)\n (-0.32783606947941524, 1.9302674946467009)\n (-0.13732511816023366, 2.141708832043786)\n (0.5089900787456267, 2.7771571126470387)\n```\n:::\n:::\n\n\nFor simple models like this one, we can call `Plots.plot` on our instance, fit result and data to generate the chart below:\n\n::: {.cell execution_count=6}\n``` {.julia .cell-code}\nusing Plots\nzoom = -0.5\nplt = plot(mach.model, mach.fitresult, Xtest, ytest, zoom=zoom, observed_lab=\"Test points\")\nxrange = range(-xmax+zoom,xmax-zoom,length=N)\nplot!(plt, xrange, @.(fun(xrange)), lw=1, ls=:dash, colour=:black, label=\"Ground truth\")\n```\n\n::: {.cell-output .cell-output-display execution_count=31}\n{}\n:::\n:::\n\n\nWe can evaluate the conformal model using the standard [MLJ](https://alan-turing-institute.github.io/MLJ.jl/dev/) workflow with a custom performance measure. You can use either `emp_coverage` for the overall empirical coverage (correctness) or `ssc` for the size-stratified coverage rate (adaptiveness).\n\n::: {.cell execution_count=7}\n``` {.julia .cell-code}\n_eval = evaluate!(mach; measure=[emp_coverage, ssc], verbosity=0)\ndisplay(_eval)\nprintln(\"Empirical coverage: $(round(_eval.measurement[1], digits=3))\")\nprintln(\"SSC: $(round(_eval.measurement[2], digits=3))\")\n```\n\n::: {.cell-output .cell-output-display}\n```\nPerformanceEvaluation object with these fields:\n measure, operation, measurement, per_fold,\n per_observation, fitted_params_per_fold,\n report_per_fold, train_test_rows\nExtract:\n┌───────────────────────────────────────────────────────────┬───────────┬───────\n│ measure │ operation │ meas ⋯\n├───────────────────────────────────────────────────────────┼───────────┼───────\n│ emp_coverage (generic function with 1 method) │ predict │ 0.94 ⋯\n│ size_stratified_coverage (generic function with 1 method) │ predict │ 0.76 ⋯\n└───────────────────────────────────────────────────────────┴───────────┴───────\n 3 columns omitted\n```\n:::\n\n::: {.cell-output .cell-output-stdout}\n```\nEmpirical coverage: 0.947\nSSC: 0.762\n```\n:::\n:::\n\n\n## 🔁 Status \n\nThis package is in its early stages of development and therefore still subject to changes to the core architecture and API. \n\n### Implemented Methodologies\n\nThe following CP approaches have been implemented:\n\n**Regression**:\n\n- Inductive \n- Naive Transductive \n- Jackknife \n- Jackknife+ \n- Jackknife-minmax\n- CV+\n- CV-minmax\n\n**Classification**:\n\n- Inductive (LABEL [@sadinle2019least])\n- Naive Transductive \n- Adaptive Inductive\n\nThe package has been tested for the following supervised models offered by [MLJ](https://alan-turing-institute.github.io/MLJ.jl/dev/).\n\n**Regression**:\n\n::: {.cell execution_count=8}\n``` {.julia .cell-code}\nkeys(tested_atomic_models[:regression])\n```\n\n::: {.cell-output .cell-output-display execution_count=33}\n```\nKeySet for a Dict{Symbol, Expr} with 5 entries. Keys:\n :nearest_neighbor\n :evo_tree\n :light_gbm\n :linear\n :decision_tree\n```\n:::\n:::\n\n\n**Classification**:\n\n::: {.cell execution_count=9}\n``` {.julia .cell-code}\nkeys(tested_atomic_models[:classification])\n```\n\n::: {.cell-output .cell-output-display execution_count=34}\n```\nKeySet for a Dict{Symbol, Expr} with 5 entries. Keys:\n :nearest_neighbor\n :evo_tree\n :light_gbm\n :decision_tree\n :logistic\n```\n:::\n:::\n\n\n### Implemented Evaluation Metrics\n\nTo evaluate conformal predictors we are typically interested in correctness and adaptiveness. The former can be evaluated by looking at the empirical coverage rate, while the latter can be assessed through metrics that address the conditional coverage [@angelopoulos2021gentle]. To this end, the following metrics have been implemented:\n\n- `emp_coverage` (empirical coverage)\n- `ssc` (size-stratified coverage)\n\nThere is also a simple `Plots.jl` recipe that can be used to inspect the set sizes. In the regression case, the interval width is stratified into discrete bins for this purpose:\n\n::: {.cell execution_count=10}\n``` {.julia .cell-code}\nbar(mach.model, mach.fitresult, X)\n```\n\n::: {.cell-output .cell-output-display execution_count=35}\n{}\n:::\n:::\n\n\n## 🛠 Contribute \n\nContributions are welcome! A good place to start is the [list](https://github.com/pat-alt/ConformalPrediction.jl/issues) of outstanding issues. For more details, see also the [Contributor's Guide](https://www.paltmeyer.com/ConformalPrediction.jl/dev/contribute/). Please follow the [SciML ColPrac guide](https://github.com/SciML/ColPrac).\n\n## 🙏 Thanks\n\nTo build this package we have made heavy use of this amazing [tutorial](https://arxiv.org/abs/2107.07511) [@angelopoulos2021gentle] and also this research [paper](https://arxiv.org/abs/1905.02928). The Awesome Conformal Prediction [repository](https://github.com/valeman/awesome-conformal-prediction) [@manokhin2022awesome] has also been a fantastic place to get started. Special thanks also to [\\@aangelopoulos](https://github.com/aangelopoulos), [\\@valeman](https://github.com/valeman) and others for actively contributing to discussions on here. \n\n## 🎓 References \n\n",
+ "markdown": "```@meta\nCurrentModule = ConformalPrediction\n```\n\n# ConformalPrediction\n\nDocumentation for [ConformalPrediction.jl](https://github.com/pat-alt/ConformalPrediction.jl).\n\n\n\n`ConformalPrediction.jl` is a package for Uncertainty Quantification (UQ) through Conformal Prediction (CP) in Julia. It is designed to work with supervised models trained in [MLJ](https://alan-turing-institute.github.io/MLJ.jl/dev/) [@blaom2020mlj]. Conformal Prediction is distribution-free, easy-to-understand, easy-to-use and model-agnostic. \n\n## 🏃 Quick Tour\n\n> First time here? Take a quick interactive [tour](https://binder.plutojl.org/v0.19.12/open?url=https%253A%252F%252Fraw.githubusercontent.com%252Fpat-alt%252FConformalPrediction.jl%252Fmain%252Fdev%252Fquick_tour%252Fnotebook.jl) to see what this package can do. \n\nThe [link](https://binder.plutojl.org/v0.19.12/open?url=https%253A%252F%252Fraw.githubusercontent.com%252Fpat-alt%252FConformalPrediction.jl%252Fmain%252Fdev%252Fquick_tour%252Fnotebook.jl) takes you to a `Pluto.jl` notebook hosted on binder.org. In my own experience, this may take a while to load, certainly enough time to get yourself a hot beverage ☕. Alternatively, you can clone this repo and run the notebook contained in `dev/quick-tour` locally or skip the tour for now and read on here.\n\n## 📖 Background\n\nConformal Prediction is a scalable frequentist approach to uncertainty quantification and coverage control. It promises to be an easy-to-understand, distribution-free and model-agnostic way to generate statistically rigorous uncertainty estimates. Interestingly, it can even be used to complement Bayesian methods.\n\nThe animation below is lifted from a small blog post that introduces the topic and the package ([[TDS](https://towardsdatascience.com/conformal-prediction-in-julia-351b81309e30)], [[Quarto](https://www.paltmeyer.com/blog/posts/conformal-prediction/#fig-anim)]). It shows conformal prediction sets for two different samples and changing coverage rates. Standard conformal classifiers produce set-valued predictions: for ambiguous samples these sets are typically large (for high coverage) or empty (for low coverage).\n\n\n\n## 🚩 Installation \n\nYou can install the latest stable release from the general registry:\n\n```{.julia}\nusing Pkg\nPkg.add(\"ConformalPrediction\")\n```\n\nThe development version can be installed as follows:\n\n```{.julia}\nusing Pkg\nPkg.add(url=\"https://github.com/pat-alt/ConformalPrediction.jl\")\n```\n\n## 🔍 Usage Example \n\nTo illustrate the intended use of the package, let's have a quick look at a simple regression problem. We first generate some synthetic data and then determine indices for our training and test data using [MLJ](https://alan-turing-institute.github.io/MLJ.jl/dev/):\n\n::: {.cell execution_count=2}\n``` {.julia .cell-code}\nusing MLJ\n\n# Inputs:\nN = 600\nxmax = 3.0\nusing Distributions\nd = Uniform(-xmax, xmax)\nX = rand(d, N)\nX = reshape(X, :, 1)\n\n# Outputs:\nnoise = 0.5\nfun(X) = X * sin(X)\nε = randn(N) .* noise\ny = @.(fun(X)) + ε\ny = vec(y)\n\n# Partition:\ntrain, test = partition(eachindex(y), 0.4, 0.4, shuffle=true)\n```\n:::\n\n\nWe then import a decision-tree based regressor ([`EvoTrees.jl`](https://github.com/Evovest/EvoTrees.jl)) following the standard [MLJ](https://alan-turing-institute.github.io/MLJ.jl/dev/) procedure.\n\n::: {.cell execution_count=3}\n``` {.julia .cell-code}\nEvoTreeRegressor = @load EvoTreeRegressor pkg=EvoTrees\nmodel = EvoTreeRegressor(rounds=100) \n```\n:::\n\n\nTo turn our conventional model into a conformal model, we just need to declare it as such by using `conformal_model` wrapper function. The generated conformal model instance can wrapped in data to create a *machine*. Finally, we proceed by fitting the machine on training data using the generic `fit!` method:\n\n::: {.cell execution_count=4}\n``` {.julia .cell-code}\nusing ConformalPrediction\nconf_model = conformal_model(model; method=:jackknife_plus)\nmach = machine(conf_model, X, y)\nfit!(mach, rows=train)\n```\n:::\n\n\nPredictions can then be computed using the generic `predict` method. The code below produces predictions for the first `n` samples. Each tuple contains the lower and upper bound for the prediction interval. \n\n::: {.cell execution_count=5}\n``` {.julia .cell-code}\nshow_first = 5\nXtest = selectrows(X, test)\nytest = y[test]\nŷ = predict(mach, Xtest)\nŷ[1:show_first]\n```\n\n::: {.cell-output .cell-output-display execution_count=6}\n```\n5-element Vector{Tuple{Float64, Float64}}:\n (0.325476135568554, 2.5420611849529986)\n (-0.8093221495344456, 1.513229072277355)\n (0.24246467414510378, 2.531848511672005)\n (-0.37629465789570005, 1.9144457517084361)\n (-0.5411423339519135, 1.712803571302072)\n```\n:::\n:::\n\n\nFor simple models like this one, we can call a custom `Plots` recipe on our instance, fit result and data to generate the chart below:\n\n::: {.cell execution_count=6}\n``` {.julia .cell-code}\nusing Plots\nzoom = -0.5\nplt = plot(mach.model, mach.fitresult, Xtest, ytest, zoom=zoom, observed_lab=\"Test points\")\nxrange = range(-xmax+zoom,xmax-zoom,length=N)\nplot!(plt, xrange, @.(fun(xrange)), lw=1, ls=:dash, colour=:black, label=\"Ground truth\")\n```\n\n::: {.cell-output .cell-output-display execution_count=7}\n{}\n:::\n:::\n\n\nWe can evaluate the conformal model using the standard [MLJ](https://alan-turing-institute.github.io/MLJ.jl/dev/) workflow with a custom performance measure. You can use either `emp_coverage` for the overall empirical coverage (correctness) or `ssc` for the size-stratified coverage rate (adaptiveness).\n\n::: {.cell execution_count=7}\n``` {.julia .cell-code}\n_eval = evaluate!(mach; measure=[emp_coverage, ssc], verbosity=0)\ndisplay(_eval)\nprintln(\"Empirical coverage: $(round(_eval.measurement[1], digits=3))\")\nprintln(\"SSC: $(round(_eval.measurement[2], digits=3))\")\n```\n\n::: {.cell-output .cell-output-display}\n```\nPerformanceEvaluation object with these fields:\n measure, operation, measurement, per_fold,\n per_observation, fitted_params_per_fold,\n report_per_fold, train_test_rows\nExtract:\n┌───────────────────────────────────────────────────────────┬───────────┬───────\n│ measure │ operation │ meas ⋯\n├───────────────────────────────────────────────────────────┼───────────┼───────\n│ emp_coverage (generic function with 1 method) │ predict │ 0.95 ⋯\n│ size_stratified_coverage (generic function with 1 method) │ predict │ 0.90 ⋯\n└───────────────────────────────────────────────────────────┴───────────┴───────\n 3 columns omitted\n```\n:::\n\n::: {.cell-output .cell-output-stdout}\n```\nEmpirical coverage: 0.957\nSSC: 0.907\n```\n:::\n:::\n\n\n## 🔁 Status \n\nThis package is in its early stages of development and therefore still subject to changes to the core architecture and API. \n\n### Implemented Methodologies\n\nThe following CP approaches have been implemented:\n\n**Regression**:\n\n- Inductive \n- Naive Transductive \n- Jackknife \n- Jackknife+ \n- Jackknife-minmax\n- CV+\n- CV-minmax\n\n**Classification**:\n\n- Inductive (LABEL [@sadinle2019least])\n- Naive Transductive \n- Adaptive Inductive\n\nThe package has been tested for the following supervised models offered by [MLJ](https://alan-turing-institute.github.io/MLJ.jl/dev/).\n\n**Regression**:\n\n::: {.cell execution_count=8}\n``` {.julia .cell-code}\nkeys(tested_atomic_models[:regression])\n```\n\n::: {.cell-output .cell-output-display execution_count=9}\n```\nKeySet for a Dict{Symbol, Expr} with 5 entries. Keys:\n :nearest_neighbor\n :evo_tree\n :light_gbm\n :linear\n :decision_tree\n```\n:::\n:::\n\n\n**Classification**:\n\n::: {.cell execution_count=9}\n``` {.julia .cell-code}\nkeys(tested_atomic_models[:classification])\n```\n\n::: {.cell-output .cell-output-display execution_count=10}\n```\nKeySet for a Dict{Symbol, Expr} with 5 entries. Keys:\n :nearest_neighbor\n :evo_tree\n :light_gbm\n :decision_tree\n :logistic\n```\n:::\n:::\n\n\n### Implemented Evaluation Metrics\n\nTo evaluate conformal predictors we are typically interested in correctness and adaptiveness. The former can be evaluated by looking at the empirical coverage rate, while the latter can be assessed through metrics that address the conditional coverage [@angelopoulos2021gentle]. To this end, the following metrics have been implemented:\n\n- `emp_coverage` (empirical coverage)\n- `ssc` (size-stratified coverage)\n\nThere is also a simple `Plots.jl` recipe that can be used to inspect the set sizes. In the regression case, the interval width is stratified into discrete bins for this purpose:\n\n::: {.cell execution_count=10}\n``` {.julia .cell-code}\nbar(mach.model, mach.fitresult, X)\n```\n\n::: {.cell-output .cell-output-display execution_count=11}\n{}\n:::\n:::\n\n\n## 🛠 Contribute \n\nContributions are welcome! A good place to start is the [list](https://github.com/pat-alt/ConformalPrediction.jl/issues) of outstanding issues. For more details, see also the [Contributor's Guide](https://www.paltmeyer.com/ConformalPrediction.jl/dev/contribute/). Please follow the [SciML ColPrac guide](https://github.com/SciML/ColPrac).\n\n## 🙏 Thanks\n\nTo build this package we have made heavy use of this amazing [tutorial](https://arxiv.org/abs/2107.07511) [@angelopoulos2021gentle] and also this research [paper](https://arxiv.org/abs/1905.02928). The Awesome Conformal Prediction [repository](https://github.com/valeman/awesome-conformal-prediction) [@manokhin2022awesome] has also been a fantastic place to get started. Special thanks also to [\\@aangelopoulos](https://github.com/aangelopoulos), [\\@valeman](https://github.com/valeman) and others for actively contributing to discussions on here. \n\n## 🎓 References \n\n",
"supporting": [
"index_files"
],
diff --git a/_freeze/docs/src/index/figure-commonmark/cell-11-output-1.svg b/_freeze/docs/src/index/figure-commonmark/cell-11-output-1.svg
index 2548e06..cb59e36 100644
--- a/_freeze/docs/src/index/figure-commonmark/cell-11-output-1.svg
+++ b/_freeze/docs/src/index/figure-commonmark/cell-11-output-1.svg
@@ -1,125 +1,119 @@
diff --git a/_freeze/docs/src/index/figure-commonmark/cell-7-output-1.svg b/_freeze/docs/src/index/figure-commonmark/cell-7-output-1.svg
index f12ab78..e543295 100644
--- a/_freeze/docs/src/index/figure-commonmark/cell-7-output-1.svg
+++ b/_freeze/docs/src/index/figure-commonmark/cell-7-output-1.svg
@@ -1,439 +1,496 @@
+
diff --git a/docs/src/_intro.qmd b/docs/src/_intro.qmd
index 0a181f4..81d3a1c 100644
--- a/docs/src/_intro.qmd
+++ b/docs/src/_intro.qmd
@@ -10,7 +10,13 @@ using ConformalPrediction
`ConformalPrediction.jl` is a package for Uncertainty Quantification (UQ) through Conformal Prediction (CP) in Julia. It is designed to work with supervised models trained in [MLJ](https://alan-turing-institute.github.io/MLJ.jl/dev/) [@blaom2020mlj]. Conformal Prediction is distribution-free, easy-to-understand, easy-to-use and model-agnostic.
-# 📖 Background
+## 🏃 Quick Tour
+
+> First time here? Take a quick interactive [tour](https://binder.plutojl.org/v0.19.12/open?url=https%253A%252F%252Fraw.githubusercontent.com%252Fpat-alt%252FConformalPrediction.jl%252Fmain%252Fdev%252Fquick_tour%252Fnotebook.jl) to see what this package can do.
+
+The [link](https://binder.plutojl.org/v0.19.12/open?url=https%253A%252F%252Fraw.githubusercontent.com%252Fpat-alt%252FConformalPrediction.jl%252Fmain%252Fdev%252Fquick_tour%252Fnotebook.jl) takes you to a `Pluto.jl` notebook hosted on binder.org. In my own experience, this may take a while to load, certainly enough time to get yourself a hot beverage ☕. Alternatively, you can clone this repo and run the notebook contained in `dev/quick-tour` locally or skip the tour for now and read on here.
+
+## 📖 Background
Conformal Prediction is a scalable frequentist approach to uncertainty quantification and coverage control. It promises to be an easy-to-understand, distribution-free and model-agnostic way to generate statistically rigorous uncertainty estimates. Interestingly, it can even be used to complement Bayesian methods.
diff --git a/docs/src/index.md b/docs/src/index.md
index fd50000..6a10e10 100644
--- a/docs/src/index.md
+++ b/docs/src/index.md
@@ -9,7 +9,13 @@ Documentation for [ConformalPrediction.jl](https://github.com/pat-alt/ConformalP
`ConformalPrediction.jl` is a package for Uncertainty Quantification (UQ) through Conformal Prediction (CP) in Julia. It is designed to work with supervised models trained in [MLJ](https://alan-turing-institute.github.io/MLJ.jl/dev/) (Blaom et al. 2020). Conformal Prediction is distribution-free, easy-to-understand, easy-to-use and model-agnostic.
-# 📖 Background
+## 🏃 Quick Tour
+
+> First time here? Take a quick interactive [tour](https://binder.plutojl.org/v0.19.12/open?url=https%253A%252F%252Fraw.githubusercontent.com%252Fpat-alt%252FConformalPrediction.jl%252Fmain%252Fdev%252Fquick_tour%252Fnotebook.jl) to see what this package can do.
+
+The [link](https://binder.plutojl.org/v0.19.12/open?url=https%253A%252F%252Fraw.githubusercontent.com%252Fpat-alt%252FConformalPrediction.jl%252Fmain%252Fdev%252Fquick_tour%252Fnotebook.jl) takes you to a `Pluto.jl` notebook hosted on binder.org. In my own experience, this may take a while to load, certainly enough time to get yourself a hot beverage ☕. Alternatively, you can clone this repo and run the notebook contained in `dev/quick-tour` locally or skip the tour for now and read on here.
+
+## 📖 Background
Conformal Prediction is a scalable frequentist approach to uncertainty quantification and coverage control. It promises to be an easy-to-understand, distribution-free and model-agnostic way to generate statistically rigorous uncertainty estimates. Interestingly, it can even be used to complement Bayesian methods.
@@ -86,13 +92,13 @@ ŷ[1:show_first]
```
5-element Vector{Tuple{Float64, Float64}}:
- (0.5113539995719073, 2.7791173590180245)
- (0.15501260477711076, 2.491986075800726)
- (-0.32783606947941524, 1.9302674946467009)
- (-0.13732511816023366, 2.141708832043786)
- (0.5089900787456267, 2.7771571126470387)
+ (0.325476135568554, 2.5420611849529986)
+ (-0.8093221495344456, 1.513229072277355)
+ (0.24246467414510378, 2.531848511672005)
+ (-0.37629465789570005, 1.9144457517084361)
+ (-0.5411423339519135, 1.712803571302072)
-For simple models like this one, we can call `Plots.plot` on our instance, fit result and data to generate the chart below:
+For simple models like this one, we can call a custom `Plots` recipe on our instance, fit result and data to generate the chart below:
``` julia
using Plots
@@ -121,13 +127,13 @@ println("SSC: $(round(_eval.measurement[2], digits=3))")
┌───────────────────────────────────────────────────────────┬───────────┬───────
│ measure │ operation │ meas ⋯
├───────────────────────────────────────────────────────────┼───────────┼───────
- │ emp_coverage (generic function with 1 method) │ predict │ 0.94 ⋯
- │ size_stratified_coverage (generic function with 1 method) │ predict │ 0.76 ⋯
+ │ emp_coverage (generic function with 1 method) │ predict │ 0.95 ⋯
+ │ size_stratified_coverage (generic function with 1 method) │ predict │ 0.90 ⋯
└───────────────────────────────────────────────────────────┴───────────┴───────
3 columns omitted
- Empirical coverage: 0.947
- SSC: 0.762
+ Empirical coverage: 0.957
+ SSC: 0.907
## 🔁 Status
diff --git a/docs/src/index_files/figure-commonmark/cell-11-output-1.svg b/docs/src/index_files/figure-commonmark/cell-11-output-1.svg
index 2548e06..cb59e36 100644
--- a/docs/src/index_files/figure-commonmark/cell-11-output-1.svg
+++ b/docs/src/index_files/figure-commonmark/cell-11-output-1.svg
@@ -1,125 +1,119 @@
diff --git a/docs/src/index_files/figure-commonmark/cell-7-output-1.svg b/docs/src/index_files/figure-commonmark/cell-7-output-1.svg
index f12ab78..e543295 100644
--- a/docs/src/index_files/figure-commonmark/cell-7-output-1.svg
+++ b/docs/src/index_files/figure-commonmark/cell-7-output-1.svg
@@ -1,439 +1,496 @@
+
diff --git a/test/Manifest.toml b/test/Manifest.toml
index 13bdb06..05d4057 100644
--- a/test/Manifest.toml
+++ b/test/Manifest.toml
@@ -2,7 +2,7 @@
julia_version = "1.8.1"
manifest_format = "2.0"
-project_hash = "c86d41ede7b316f1c0c615053739e4cfe0ac765b"
+project_hash = "927d1b7a82bc93ab3d48f603d6d16919bd6f0dc1"
[[deps.ANSIColoredPrinters]]
git-tree-sha1 = "574baf8110975760d391c710b6341da1afa48d8c"
@@ -216,12 +216,6 @@ git-tree-sha1 = "52cb3ec90e8a8bea0e62e275ba577ad0f74821f7"
uuid = "ed09eef8-17a6-5b46-8889-db040fac31e3"
version = "0.3.2"
-[[deps.Conda]]
-deps = ["Downloads", "JSON", "VersionParsing"]
-git-tree-sha1 = "6e47d11ea2776bc5627421d59cdcc1296c058071"
-uuid = "8f4d0f93-b110-5947-807f-2305c1781a2d"
-version = "1.7.0"
-
[[deps.ConstructionBase]]
deps = ["LinearAlgebra"]
git-tree-sha1 = "fb21ddd70a051d882a1686a5a550990bbe371a95"
@@ -249,12 +243,6 @@ git-tree-sha1 = "e08915633fcb3ea83bf9d6126292e5bc5c739922"
uuid = "9a962f9c-6df0-11e9-0e5d-c546b8b5ee8a"
version = "1.13.0"
-[[deps.DataFrames]]
-deps = ["Compat", "DataAPI", "Future", "InvertedIndices", "IteratorInterfaceExtensions", "LinearAlgebra", "Markdown", "Missings", "PooledArrays", "PrettyTables", "Printf", "REPL", "Random", "Reexport", "SnoopPrecompile", "SortingAlgorithms", "Statistics", "TableTraits", "Tables", "Unicode"]
-git-tree-sha1 = "d4f69885afa5e6149d0cab3818491565cf41446d"
-uuid = "a93c6f00-e57d-5684-b7b6-d8193f3e46c0"
-version = "1.4.4"
-
[[deps.DataStructures]]
deps = ["Compat", "InteractiveUtils", "OrderedCollections"]
git-tree-sha1 = "d1fff3a548102f48987a52a2e0d114fa97d730f0"
@@ -851,12 +839,6 @@ git-tree-sha1 = "08203fc87a7f992cee24e7a1b2353e594c73c41c"
uuid = "d491faf4-2d78-11e9-2867-c94bc002c0b7"
version = "0.16.2"
-[[deps.MLJScikitLearnInterface]]
-deps = ["MLJModelInterface", "PyCall", "ScikitLearn"]
-git-tree-sha1 = "d8e900e2676df6b9427063f5ad39761dce5e5761"
-uuid = "5ae90465-5518-4432-b9d2-8a1def2f0cab"
-version = "0.2.0"
-
[[deps.MLJTuning]]
deps = ["ComputationalResources", "Distributed", "Distributions", "LatinHypercubeSampling", "MLJBase", "ProgressMeter", "Random", "RecipesBase"]
git-tree-sha1 = "02688098bd77827b64ed8ad747c14f715f98cfc4"
@@ -1073,12 +1055,6 @@ git-tree-sha1 = "050ca4aa2ca31484b51b849d8180caf8e4449c49"
uuid = "1d0040c9-8b98-4ee7-8388-3f51789ca0ad"
version = "0.1.11"
-[[deps.PooledArrays]]
-deps = ["DataAPI", "Future"]
-git-tree-sha1 = "a6062fe4063cdafe78f4a0a81cfffb89721b30e7"
-uuid = "2dfb63ee-cc39-5dd5-95bd-886bf059d720"
-version = "1.4.2"
-
[[deps.PositiveFactorizations]]
deps = ["LinearAlgebra"]
git-tree-sha1 = "17275485f373e6673f7e7f97051f703ed5b15b20"
@@ -1112,12 +1088,6 @@ git-tree-sha1 = "d7a7aef8f8f2d537104f170139553b14dfe39fe9"
uuid = "92933f4c-e287-5a05-a399-4b506db050ca"
version = "1.7.2"
-[[deps.PyCall]]
-deps = ["Conda", "Dates", "Libdl", "LinearAlgebra", "MacroTools", "Serialization", "VersionParsing"]
-git-tree-sha1 = "53b8b07b721b77144a0fbbbc2675222ebf40a02d"
-uuid = "438e738f-606a-5dbb-bf0a-cddfbfd45ab0"
-version = "1.94.1"
-
[[deps.Qt5Base_jll]]
deps = ["Artifacts", "CompilerSupportLibraries_jll", "Fontconfig_jll", "Glib_jll", "JLLWrappers", "Libdl", "Libglvnd_jll", "OpenSSL_jll", "Pkg", "Xorg_libXext_jll", "Xorg_libxcb_jll", "Xorg_xcb_util_image_jll", "Xorg_xcb_util_keysyms_jll", "Xorg_xcb_util_renderutil_jll", "Xorg_xcb_util_wm_jll", "Zlib_jll", "xkbcommon_jll"]
git-tree-sha1 = "0c03844e2231e12fda4d0086fd7cbe4098ee8dc5"
@@ -1223,12 +1193,6 @@ git-tree-sha1 = "a8e18eb383b5ecf1b5e6fc237eb39255044fd92b"
uuid = "30f210dd-8aff-4c5f-94ba-8e64358c1161"
version = "3.0.0"
-[[deps.ScikitLearn]]
-deps = ["Compat", "Conda", "DataFrames", "Distributed", "IterTools", "LinearAlgebra", "MacroTools", "Parameters", "Printf", "PyCall", "Random", "ScikitLearnBase", "SparseArrays", "StatsBase", "VersionParsing"]
-git-tree-sha1 = "de6a32950c170e5fd5a2d8bcba0fb97a1028ab06"
-uuid = "3646fa90-6ef7-5e7e-9f22-8aca16db6324"
-version = "0.6.5"
-
[[deps.ScikitLearnBase]]
deps = ["LinearAlgebra", "Random", "Statistics"]
git-tree-sha1 = "7877e55c1523a4b336b433da39c8e8c08d2f221f"
@@ -1447,11 +1411,6 @@ git-tree-sha1 = "fc79d0f926592ecaeaee164f6a4ca81b51115c3b"
uuid = "3d5dd08c-fd9d-11e8-17fa-ed2836048c2f"
version = "0.21.56"
-[[deps.VersionParsing]]
-git-tree-sha1 = "58d6e80b4ee071f5efd07fda82cb9fbe17200868"
-uuid = "81def892-9a0e-5fdd-b105-ffc91e053289"
-version = "1.3.0"
-
[[deps.Wayland_jll]]
deps = ["Artifacts", "Expat_jll", "JLLWrappers", "Libdl", "Libffi_jll", "Pkg", "XML2_jll"]
git-tree-sha1 = "3e61f0b86f90dacb0bc0e73a0c5a83f6a8636e23"
diff --git a/test/Project.toml b/test/Project.toml
index 76c06f2..1872821 100644
--- a/test/Project.toml
+++ b/test/Project.toml
@@ -6,7 +6,6 @@ MLJ = "add582a8-e3ab-11e8-2d5e-e98b27df1bc7"
MLJDecisionTreeInterface = "c6f25543-311c-4c74-83dc-3ea6d1015661"
MLJLinearModels = "6ee0df7b-362f-4a72-a706-9e79364fb692"
MLJModelInterface = "e80e1ace-859a-464e-9ed9-23947d8ae3ea"
-MLJScikitLearnInterface = "5ae90465-5518-4432-b9d2-8a1def2f0cab"
NearestNeighborModels = "636a865e-7cf4-491e-846c-de09b730eb36"
Plots = "91a5bcdd-55d7-5caf-9e0b-520d859cae80"
Test = "8dfed614-e22c-5e08-85e1-65c5234f0b40"