diff --git a/.gitignore b/.gitignore
index 6ad1c578..b6706876 100644
--- a/.gitignore
+++ b/.gitignore
@@ -63,6 +63,7 @@ instance/
# Sphinx documentation
docs/_build/
+docs/output.txt
# PyBuilder
target/
@@ -108,3 +109,17 @@ info/
# Cython generated C code
*.c
+*.cpp
+
+
+# generated docs
+docs/source/examples/
+docs/source/generated/
+
+
+*.DS_Store
+
+*.zip
+
+*.vtu
+*.vtr
diff --git a/.travis.yml b/.travis.yml
index 730a6def..125eed4b 100644
--- a/.travis.yml
+++ b/.travis.yml
@@ -1,137 +1,100 @@
language: python
+python: 3.8
-matrix:
- include:
- - name: "Linux py27"
- sudo: required
- language: python
- python: 2.7
- services: docker
- env:
- - PIP=pip
- - CIBW_BUILD="cp27-*"
- - COVER="off"
+# setuptools-scm needs all tags in order to obtain a proper version
+git:
+ depth: false
+
+env:
+ global:
+ - TWINE_USERNAME=geostatframework
+ - CIBW_BEFORE_BUILD="pip install numpy==1.17.3 cython==0.29.14 setuptools"
+ - CIBW_TEST_REQUIRES=pytest
+ - CIBW_TEST_COMMAND="pytest -v {project}/tests"
- - name: "Linux py34"
- sudo: required
- language: python
- python: 3.4
+before_install:
+ - |
+ if [[ "$TRAVIS_OS_NAME" = windows ]]; then
+ choco install python --version 3.8.0
+ export PATH="/c/Python38:/c/Python38/Scripts:$PATH"
+ # make sure it's on PATH as 'python3'
+ ln -s /c/Python38/python.exe /c/Python38/python3.exe
+ fi
+
+script:
+ - python3 -m pip install cibuildwheel==1.3.0
+ - python3 -m cibuildwheel --output-dir dist
+
+after_success:
+ - |
+ if [[ $TRAVIS_PULL_REQUEST == 'false' ]]; then
+ python3 -m pip install twine
+ python3 -m twine upload --verbose --skip-existing --repository-url https://test.pypi.org/legacy/ dist/*
+ if [[ $TRAVIS_TAG ]]; then python3 -m twine upload --verbose --skip-existing dist/*; fi
+ fi
+
+notifications:
+ email:
+ recipients:
+ - info@geostat-framework.org
+
+jobs:
+ include:
+ - name: "sdist and coverage"
services: docker
- env:
- - PIP=pip
- - CIBW_BUILD="cp34-*"
- - COVER="off"
+ env: OMP_NUM_THREADS=4
+ script:
+ - python3 -m pip install -U setuptools pytest-cov coveralls
+ - python3 -m pip install -U numpy==1.17.3 cython==0.29.14
+ - python3 -m pip install -r requirements.txt
+ - python3 setup.py sdist -d dist
+ - python3 setup.py --openmp build_ext --inplace
+ - python3 -m pytest --cov gstools --cov-report term-missing -v tests/
+ - python3 -m coveralls
- name: "Linux py35"
- sudo: required
- language: python
- python: 3.5
services: docker
- env:
- - PIP=pip
- - CIBW_BUILD="cp35-*"
- - COVER="off"
-
- # py36 for coverage and sdist
+ env: CIBW_BUILD="cp35-*"
- name: "Linux py36"
- sudo: required
- language: python
- python: 3.6
services: docker
- env:
- - PIP=pip
- - CIBW_BUILD="cp36-*"
- - COVER="on"
-
- # https://github.com/travis-ci/travis-ci/issues/9815
+ env: CIBW_BUILD="cp36-*"
- name: "Linux py37"
- sudo: required
- language: python
- python: 3.7
- dist: xenial
services: docker
- env:
- - PIP=pip
- - CIBW_BUILD="cp37-*"
- - COVER="off"
-
- - name: "MacOS py27"
- os: osx
- language: generic
- env:
- - PIP=pip2
- - CIBW_BUILD="cp27-*"
- - COVER="off"
-
- - name: "MacOS py34"
- os: osx
- language: generic
- env:
- - PIP=pip2
- - CIBW_BUILD="cp34-*"
- - COVER="off"
+ env: CIBW_BUILD="cp37-*"
+ - name: "Linux py38"
+ services: docker
+ env: CIBW_BUILD="cp38-*"
- name: "MacOS py35"
os: osx
- language: generic
- env:
- - PIP=pip2
- - CIBW_BUILD="cp35-*"
- - COVER="off"
-
+ language: shell
+ env: CIBW_BUILD="cp35-*"
- name: "MacOS py36"
os: osx
- language: generic
- env:
- - PIP=pip2
- - CIBW_BUILD="cp36-*"
- - COVER="off"
-
+ language: shell
+ env: CIBW_BUILD="cp36-*"
- name: "MacOS py37"
os: osx
- language: generic
- env:
- - PIP=pip2
- - CIBW_BUILD="cp37-*"
- - COVER="off"
-
-env:
- global:
- - TWINE_USERNAME=geostatframework
- - CIBW_BEFORE_BUILD="pip install numpy==1.14.5 cython==0.28.3"
- - CIBW_TEST_REQUIRES=pytest-cov
- # inplace cython build and test run
- - CIBW_TEST_COMMAND="cd {project} && python setup.py build_ext --inplace && py.test --cov gstools --cov-report term-missing -v {project}/tests"
-
-script:
- # create wheels
- - $PIP install cibuildwheel==0.11.1
- - cibuildwheel --output-dir wheelhouse
- # create source dist for pypi and create coverage (only once for linux py3.6)
- - |
- if [[ $COVER == "on" ]]; then
- rm -rf dist
- python -m pip install -U numpy==1.14.5 cython==0.28.3 setuptools
- python -m pip install pytest-cov coveralls
- python -m pip install -r docs/requirements.txt
- python setup.py sdist
- python setup.py build_ext --inplace
- python -m pytest --cov gstools --cov-report term-missing -v tests/
- python -m coveralls
- fi
-
-after_success:
- # pypi upload ("test" allways and "official" on TAG)
- - python -m pip install twine
- - python -m twine upload --verbose --skip-existing --repository-url https://test.pypi.org/legacy/ wheelhouse/*.whl
- - python -m twine upload --verbose --skip-existing --repository-url https://test.pypi.org/legacy/ dist/*.tar.gz
- - |
- if [[ $TRAVIS_TAG ]]; then
- python -m twine upload --verbose --skip-existing wheelhouse/*.whl
- python -m twine upload --verbose --skip-existing dist/*.tar.gz
- fi
+ language: shell
+ env: CIBW_BUILD="cp37-*"
+ - name: "MacOS py38"
+ os: osx
+ language: shell
+ env: CIBW_BUILD="cp38-*"
-notifications:
- email:
- recipients:
- - info@geostat-framework.org
+ - name: "Win py35"
+ os: windows
+ language: shell
+ env: CIBW_BUILD="cp35-*"
+ - name: "Win py36"
+ os: windows
+ language: shell
+ env: CIBW_BUILD="cp36-*"
+ - name: "Win py37"
+ os: windows
+ language: shell
+ env: CIBW_BUILD="cp37-*"
+ - name: "Win py38"
+ os: windows
+ language: shell
+ env: CIBW_BUILD="cp38-*"
diff --git a/.zenodo.json b/.zenodo.json
new file mode 100755
index 00000000..ad72d74b
--- /dev/null
+++ b/.zenodo.json
@@ -0,0 +1,49 @@
+{
+ "license": "LGPL-3.0+",
+ "contributors": [
+ {
+ "type": "Other",
+ "name": "Bane Sullivan"
+ },
+ {
+ "orcid": "0000-0002-2547-8102",
+ "affiliation": "Helmholtz Centre for Environmental Research - UFZ",
+ "type": "ResearchGroup",
+ "name": "Falk He\u00dfe"
+ },
+ {
+ "orcid": "0000-0002-8783-6198",
+ "affiliation": "Hydrogeology Group, Department of Earth Science, Utrecht University, Netherlands",
+ "type": "ResearchGroup",
+ "name": "Alraune Zech"
+ },
+ {
+ "orcid": "0000-0002-7798-7080",
+ "affiliation": "Helmholtz Centre for Environmental Research - UFZ",
+ "type": "Supervisor",
+ "name": "Sabine Attinger"
+ }
+ ],
+ "language": "eng",
+ "keywords": [
+ "geostatistics",
+ "kriging",
+ "random fields",
+ "covariance models",
+ "variogram",
+ "Python",
+ "GeoStat-Framework"
+ ],
+ "creators": [
+ {
+ "orcid": "0000-0001-9060-4008",
+ "affiliation": "Helmholtz Centre for Environmental Research - UFZ",
+ "name": "Sebastian M\u00fcller"
+ },
+ {
+ "orcid": "0000-0001-9362-1372",
+ "affiliation": "Helmholtz Centre for Environmental Research - UFZ",
+ "name": "Lennart Sch\u00fcler"
+ }
+ ]
+}
\ No newline at end of file
diff --git a/CHANGELOG.md b/CHANGELOG.md
index d5885da8..8192866f 100755
--- a/CHANGELOG.md
+++ b/CHANGELOG.md
@@ -3,13 +3,30 @@
All notable changes to **GSTools** will be documented in this file.
-## [Unreleased]
+## [1.2.0] - Volatile Violet - 2020-03-20
### Enhancements
+- different variogram estimator functions can now be used #51
+- the TPLGaussian and TPLExponential now have analytical spectra #67
+- added property ``is_isotropic`` to CovModel #67
+- reworked the whole krige sub-module to provide multiple kriging methods #67
+ - Simple
+ - Ordinary
+ - Universal
+ - External Drift Kriging
+ - Detrended Kriging
+- a new transformation function for discrete fields has been added #70
+- reworked tutorial section in the documentation #63
+- pyvista interface #29
### Changes
+- Python versions 2.7 and 3.4 are no longer supported #40 #43
+- CovModel: in 3D the input of anisotropy is now treated slightly different: #67
+ - single given anisotropy value [e] is converted to [1, e] (it was [e, e] before)
+ - two given length-scales [l_1, l_2] are converted to [l_1, l_2, l_2] (it was [l_1, l_2, l_1] before)
### Bugfixes
+- a race condition in the structured variogram estimation has been fixed #51
## [1.1.1] - Reverberating Red - 2019-11-08
@@ -97,7 +114,8 @@ All notable changes to **GSTools** will be documented in this file.
First release of GSTools.
-[Unreleased]: https://github.com/GeoStat-Framework/gstools/compare/v1.1.1...HEAD
+[Unreleased]: https://github.com/GeoStat-Framework/gstools/compare/v1.2.0...HEAD
+[1.2.0]: https://github.com/GeoStat-Framework/gstools/compare/v1.1.1...v1.2.0
[1.1.1]: https://github.com/GeoStat-Framework/gstools/compare/v1.1.0...v1.1.1
[1.1.0]: https://github.com/GeoStat-Framework/gstools/compare/v1.0.1...v1.1.0
[1.0.1]: https://github.com/GeoStat-Framework/gstools/compare/v1.0.0...v1.0.1
diff --git a/CONTRIBUTING.md b/CONTRIBUTING.md
index fb253d03..efa19903 100644
--- a/CONTRIBUTING.md
+++ b/CONTRIBUTING.md
@@ -23,7 +23,7 @@ print(gstools.__version__)
Open a [new issue](https://github.com/GeoStat-Framework/GSTools/issues)
with your idea or suggestion and we'd love to discuss about it.
-
+
## Do you want to enhance GSTools or fix something?
@@ -31,4 +31,6 @@ with your idea or suggestion and we'd love to discuss about it.
- Add yourself to AUTHORS.md (if you want to).
- We use the black code format, please use the script `black --line-length 79 gstools/` after you have written your code.
- Add some tests if possible.
+- Add an example showing your new feature in one of the examples sub-folders if possible.
+ Follow this [Sphinx-Gallary guide](https://sphinx-gallery.github.io/stable/syntax.html#embed-rst-in-your-example-python-files)
- Push to your fork and submit a pull request.
diff --git a/README.md b/README.md
index 33b7aceb..24a7774a 100644
--- a/README.md
+++ b/README.md
@@ -2,10 +2,10 @@
[](https://doi.org/10.5281/zenodo.1313628)
[](https://badge.fury.io/py/gstools)
+[](https://anaconda.org/conda-forge/gstools)
[](https://travis-ci.org/GeoStat-Framework/GSTools)
-[](https://ci.appveyor.com/project/GeoStat-Framework/gstools/branch/master)
[](https://coveralls.io/github/GeoStat-Framework/GSTools?branch=master)
-[](https://geostat-framework.readthedocs.io/projects/gstools/en/latest/)
+[](https://geostat-framework.readthedocs.io/projects/gstools/en/stable/?badge=stable)
[](https://github.com/ambv/black)
@@ -58,7 +58,9 @@ To install the latest development version via pip, see the
At the moment you can cite the Zenodo code publication of GSTools:
-> Sebastian Müller, & Lennart Schüler. (2019, October 1). GeoStat-Framework/GSTools: Reverberating Red (Version v1.1.0). Zenodo. http://doi.org/10.5281/zenodo.3468230
+> Sebastian Müller & Lennart Schüler. GeoStat-Framework/GSTools. Zenodo. https://doi.org/10.5281/zenodo.1313628
+
+If you want to cite a specific version, have a look at the Zenodo site.
A publication for the GeoStat-Framework is in preperation.
@@ -79,8 +81,9 @@ The documentation also includes some [tutorials][tut_link], showing the most imp
- [Kriging][tut5_link]
- [Conditioned random field generation][tut6_link]
- [Field transformations][tut7_link]
+- [Miscellaneous examples][tut0_link]
-Some more examples are provided in the examples folder.
+The associated python scripts are provided in the `examples` folder.
## Spatial Random Field Generation
@@ -97,12 +100,11 @@ The core of this library is the generation of spatial random fields. These field
This is an example of how to generate a 2 dimensional spatial random field with a gaussian covariance model.
```python
-from gstools import SRF, Gaussian
-import matplotlib.pyplot as plt
+import gstools as gs
# structured field with a size 100x100 and a grid-size of 1x1
x = y = range(100)
-model = Gaussian(dim=2, var=1, len_scale=10)
-srf = SRF(model)
+model = gs.Gaussian(dim=2, var=1, len_scale=10)
+srf = gs.SRF(model)
srf((x, y), mesh_type='structured')
srf.plot()
```
@@ -113,12 +115,11 @@ srf.plot()
A similar example but for a three dimensional field is exported to a [VTK](https://vtk.org/) file, which can be visualized with [ParaView](https://www.paraview.org/) or [PyVista](https://docs.pyvista.org) in Python:
```python
-from gstools import SRF, Gaussian
-import matplotlib.pyplot as pt
+import gstools as gs
# structured field with a size 100x100x100 and a grid-size of 1x1x1
x = y = z = range(100)
-model = Gaussian(dim=3, var=0.6, len_scale=20)
-srf = SRF(model)
+model = gs.Gaussian(dim=3, var=0.6, len_scale=20)
+srf = gs.SRF(model)
srf((x, y, z), mesh_type='structured')
srf.vtk_export('3d_field') # Save to a VTK file for ParaView
@@ -144,18 +145,18 @@ model again.
```python
import numpy as np
-from gstools import SRF, Exponential, Stable, vario_estimate_unstructured
+import gstools as gs
# generate a synthetic field with an exponential model
x = np.random.RandomState(19970221).rand(1000) * 100.
y = np.random.RandomState(20011012).rand(1000) * 100.
-model = Exponential(dim=2, var=2, len_scale=8)
-srf = SRF(model, mean=0, seed=19970221)
+model = gs.Exponential(dim=2, var=2, len_scale=8)
+srf = gs.SRF(model, mean=0, seed=19970221)
field = srf((x, y))
# estimate the variogram of the field with 40 bins
bins = np.arange(40)
-bin_center, gamma = vario_estimate_unstructured((x, y), field, bins)
+bin_center, gamma = gs.vario_estimate_unstructured((x, y), field, bins)
# fit the variogram with a stable model. (no nugget fitted)
-fit_model = Stable(dim=2)
+fit_model = gs.Stable(dim=2)
fit_model.fit_variogram(bin_center, gamma, nugget=False)
# output
ax = fit_model.plot(x_max=40)
@@ -184,8 +185,8 @@ For better visualization, we will condition a 1d field to a few "measurements",
```python
import numpy as np
-from gstools import Gaussian, SRF
import matplotlib.pyplot as plt
+import gstools as gs
# conditions
cond_pos = [0.3, 1.9, 1.1, 3.3, 4.7]
@@ -194,8 +195,8 @@ cond_val = [0.47, 0.56, 0.74, 1.47, 1.74]
gridx = np.linspace(0.0, 15.0, 151)
# spatial random field class
-model = Gaussian(dim=1, var=0.5, len_scale=2)
-srf = SRF(model)
+model = gs.Gaussian(dim=1, var=0.5, len_scale=2)
+srf = gs.SRF(model)
srf.set_condition(cond_pos, cond_val, "ordinary")
# generate the ensemble of field realizations
@@ -223,10 +224,10 @@ Here we re-implement the Gaussian covariance model by defining just a
[correlation][cor_link] function, which takes a non-dimensional distance ``h = r/l``:
```python
-from gstools import CovModel
import numpy as np
+import gstools as gs
# use CovModel as the base-class
-class Gau(CovModel):
+class Gau(gs.CovModel):
def cor(self, h):
return np.exp(-h**2)
```
@@ -248,12 +249,11 @@ spatial vector fields can be generated.
```python
import numpy as np
-import matplotlib.pyplot as plt
-from gstools import SRF, Gaussian
+import gstools as gs
x = np.arange(100)
y = np.arange(100)
-model = Gaussian(dim=2, var=1, len_scale=10)
-srf = SRF(model, generator='VectorField')
+model = gs.Gaussian(dim=2, var=1, len_scale=10)
+srf = gs.SRF(model, generator='VectorField')
srf((x, y), mesh_type='structured', seed=19841203)
srf.plot()
```
@@ -275,10 +275,10 @@ a handy [VTK][vtk_link] export routine using the `.vtk_export()` or you could
create a VTK/PyVista dataset for use in Python with to `.to_pyvista()` method:
```python
-from gstools import SRF, Gaussian
+import gstools as gs
x = y = range(100)
-model = Gaussian(dim=2, var=1, len_scale=10)
-srf = SRF(model)
+model = gs.Gaussian(dim=2, var=1, len_scale=10)
+srf = gs.SRF(model)
srf((x, y), mesh_type='structured')
srf.vtk_export("field") # Saves to a VTK file
mesh = srf.to_pyvista() # Create a VTK/PyVista dataset in memory
@@ -288,15 +288,18 @@ mesh.plot()
Which gives a RectilinearGrid VTK file ``field.vtr`` or creates a PyVista mesh
in memory for immediate 3D plotting in Python.
+
+ +
+
+
## Requirements:
- [NumPy >= 1.14.5](https://www.numpy.org)
- [SciPy >= 1.1.0](https://www.scipy.org/scipylib)
-- [hankel >= 0.3.6](https://github.com/steven-murray/hankel)
+- [hankel >= 1.0.2](https://github.com/steven-murray/hankel)
- [emcee >= 3.0.0](https://github.com/dfm/emcee)
-- [pyevtk](https://bitbucket.org/pauloh/pyevtk)
-- [six](https://github.com/benjaminp/six)
+- [pyevtk >= 1.1.1](https://github.com/pyscience-projects/pyevtk)
### Optional
@@ -311,7 +314,7 @@ You can contact us via .
## License
-[LGPLv3][license_link] © 2018-2019
+[LGPLv3][license_link] © 2018-2020
[pip_link]: https://pypi.org/project/gstools
[conda_link]: https://docs.conda.io/en/latest/miniconda.html
@@ -320,17 +323,18 @@ You can contact us via .
[pipiflag]: https://pip-python3.readthedocs.io/en/latest/reference/pip_install.html?highlight=i#cmdoption-i
[winpy_link]: https://winpython.github.io/
[license_link]: https://github.com/GeoStat-Framework/GSTools/blob/master/LICENSE
-[cov_link]: https://geostat-framework.readthedocs.io/projects/gstools/en/latest/covmodel.html
+[cov_link]: https://geostat-framework.readthedocs.io/projects/gstools/en/stable/generated/gstools.covmodel.CovModel.html#gstools.covmodel.CovModel
[stable_link]: https://en.wikipedia.org/wiki/Stable_distribution
-[doc_link]: https://geostat-framework.readthedocs.io/projects/gstools/en/latest/
-[doc_install_link]: https://geostat-framework.readthedocs.io/projects/gstools/en/latest/#pip
-[tut_link]: https://geostat-framework.readthedocs.io/projects/gstools/en/latest/tutorials.html
-[tut1_link]: https://geostat-framework.readthedocs.io/projects/gstools/en/latest/examples/01_random_field/index.html
-[tut2_link]: https://geostat-framework.readthedocs.io/projects/gstools/en/latest/examples/02_cov_model/index.html
-[tut3_link]: https://geostat-framework.readthedocs.io/projects/gstools/en/latest/examples/03_variogram/index.html
-[tut4_link]: https://geostat-framework.readthedocs.io/projects/gstools/en/latest/examples/04_vector_field/index.html
-[tut5_link]: https://geostat-framework.readthedocs.io/projects/gstools/en/latest/examples/05_kriging/index.html
-[tut6_link]: https://geostat-framework.readthedocs.io/projects/gstools/en/latest/examples/06_conditioned_fields/index.html
-[tut7_link]: https://geostat-framework.readthedocs.io/projects/gstools/en/latest/examples/07_transformations/index.html
+[doc_link]: https://geostat-framework.readthedocs.io/projects/gstools/en/stable/
+[doc_install_link]: https://geostat-framework.readthedocs.io/projects/gstools/en/stable/#pip
+[tut_link]: https://geostat-framework.readthedocs.io/projects/gstools/en/stable/tutorials.html
+[tut1_link]: https://geostat-framework.readthedocs.io/projects/gstools/en/stable/examples/01_random_field/index.html
+[tut2_link]: https://geostat-framework.readthedocs.io/projects/gstools/en/stable/examples/02_cov_model/index.html
+[tut3_link]: https://geostat-framework.readthedocs.io/projects/gstools/en/stable/examples/03_variogram/index.html
+[tut4_link]: https://geostat-framework.readthedocs.io/projects/gstools/en/stable/examples/04_vector_field/index.html
+[tut5_link]: https://geostat-framework.readthedocs.io/projects/gstools/en/stable/examples/05_kriging/index.html
+[tut6_link]: https://geostat-framework.readthedocs.io/projects/gstools/en/stable/examples/06_conditioned_fields/index.html
+[tut7_link]: https://geostat-framework.readthedocs.io/projects/gstools/en/stable/examples/07_transformations/index.html
+[tut0_link]: https://geostat-framework.readthedocs.io/projects/gstools/en/stable/examples/00_misc/index.html
[cor_link]: https://en.wikipedia.org/wiki/Autocovariance#Normalization
[vtk_link]: https://www.vtk.org/
diff --git a/appveyor.yml b/appveyor.yml
deleted file mode 100644
index acd9e40d..00000000
--- a/appveyor.yml
+++ /dev/null
@@ -1,22 +0,0 @@
-environment:
- global:
- TWINE_USERNAME: geostatframework
- CIBW_BEFORE_BUILD: pip install numpy==1.14.5 cython==0.28.3
- CIBW_TEST_REQUIRES: pytest-cov
- CIBW_TEST_COMMAND: cd {project} && python setup.py build_ext --inplace && py.test --cov gstools --cov-report term-missing -v {project}/tests
-
-build_script:
- - pip install cibuildwheel==0.11.1
- - cibuildwheel --output-dir wheelhouse
- - python -m pip install twine
- - python -m twine upload --skip-existing --repository-url https://test.pypi.org/legacy/ wheelhouse/*.whl
- - >
- IF "%APPVEYOR_REPO_TAG%" == "true"
- (
- python -m pip install twine
- &&
- python -m twine upload --skip-existing wheelhouse/*.whl
- )
-artifacts:
- - path: "wheelhouse\\*.whl"
- name: Wheels
\ No newline at end of file
diff --git a/docs/output.txt b/docs/output.txt
deleted file mode 100644
index 869370c5..00000000
--- a/docs/output.txt
+++ /dev/null
@@ -1,59 +0,0 @@
-Sphinx v1.8.2 in Verwendung
-Erstelle Ausgabeverzeichnis…
-loading intersphinx inventory from https://docs.python.org/3.6/objects.inv...
-loading intersphinx inventory from https://docs.python.org/objects.inv...
-intersphinx inventory has moved: https://docs.python.org/objects.inv -> https://docs.python.org/3/objects.inv
-loading intersphinx inventory from http://docs.scipy.org/doc/numpy/objects.inv...
-intersphinx inventory has moved: http://docs.scipy.org/doc/numpy/objects.inv -> https://docs.scipy.org/doc/numpy/objects.inv
-loading intersphinx inventory from http://docs.scipy.org/doc/scipy/reference/objects.inv...
-intersphinx inventory has moved: http://docs.scipy.org/doc/scipy/reference/objects.inv -> https://docs.scipy.org/doc/scipy/reference/objects.inv
-loading intersphinx inventory from http://matplotlib.org/objects.inv...
-intersphinx inventory has moved: http://matplotlib.org/objects.inv -> https://matplotlib.org/objects.inv
-loading intersphinx inventory from http://www.sphinx-doc.org/en/stable/objects.inv...
-[autosummary] generating autosummary for: covmodel.rst, field.rst, index.rst, main.rst, random.rst, upscaling.rst, variogram.rst
-building [mo]: targets for 0 po files that are out of date
-building [html]: targets for 7 source files that are out of date
-updating environment: 7 added, 0 changed, 0 removed
-reading sources... [ 14%] covmodel
-reading sources... [ 28%] field
-reading sources... [ 42%] index
-reading sources... [ 57%] main
-reading sources... [ 71%] random
-reading sources... [ 85%] upscaling
-reading sources... [100%] variogram
-
-looking for now-outdated files... none found
-pickling environment... erledigt
-checking consistency... erledigt
-preparing documents... erledigt
-writing output... [ 14%] covmodel
-writing output... [ 28%] field
-writing output... [ 42%] index
-writing output... [ 57%] main
-writing output... [ 71%] random
-writing output... [ 85%] upscaling
-writing output... [100%] variogram
-
-generating indices... genindex py-modindex
-highlighting module code... [ 12%] gstools.covmodel.base
-highlighting module code... [ 25%] gstools.covmodel.models
-highlighting module code... [ 37%] gstools.field.generator
-highlighting module code... [ 50%] gstools.field.srf
-highlighting module code... [ 62%] gstools.field.upscaling
-highlighting module code... [ 75%] gstools.random.rng
-highlighting module code... [ 87%] gstools.random.tools
-highlighting module code... [100%] gstools.variogram.variogram
-
-writing additional pages... search
-copying images... [ 25%] gstools.png
-copying images... [ 50%] gau_field.png
-copying images... [ 75%] tplstable_field.png
-copying images... [100%] exp_vario_fit.png
-
-copying static files... done
-copying extra files... erledigt
-dumping search index in English (code: en) ... erledigt
-dumping object inventory... erledigt
-build abgeschlossen, 5640 warnings.
-
-The HTML pages are in build/html.
diff --git a/docs/requirements.txt b/docs/requirements.txt
index 3eb3c028..c5a6a232 100644
--- a/docs/requirements.txt
+++ b/docs/requirements.txt
@@ -1,5 +1,3 @@
-# required for readthedocs.org
-cython>=0.28.3
-numpydoc
-# https://stackoverflow.com/a/11704396/6696397
--r ../requirements.txt
\ No newline at end of file
+-r requirements_doc.txt
+-r ../requirements_setup.txt
+-r ../requirements.txt
diff --git a/docs/requirements_doc.txt b/docs/requirements_doc.txt
new file mode 100755
index 00000000..c9d3ee24
--- /dev/null
+++ b/docs/requirements_doc.txt
@@ -0,0 +1,5 @@
+numpydoc
+sphinx-gallery
+matplotlib
+pyvista
+pykrige
diff --git a/docs/source/covmodel.base.rst b/docs/source/_templates/autosummary/class.rst
similarity index 53%
rename from docs/source/covmodel.base.rst
rename to docs/source/_templates/autosummary/class.rst
index 7aa1c973..c5c858a1 100644
--- a/docs/source/covmodel.base.rst
+++ b/docs/source/_templates/autosummary/class.rst
@@ -1,11 +1,12 @@
-gstools.covmodel.base
----------------------
+{{ fullname | escape | underline}}
-.. automodule:: gstools.covmodel.base
+.. currentmodule:: {{ module }}
+
+.. autoclass:: {{ objname }}
:members:
:undoc-members:
- :show-inheritance:
:inherited-members:
+ :show-inheritance:
.. raw:: latex
diff --git a/docs/source/conf.py b/docs/source/conf.py
index 1961b10a..b9214669 100644
--- a/docs/source/conf.py
+++ b/docs/source/conf.py
@@ -20,8 +20,16 @@
# NOTE:
# pip install sphinx_rtd_theme
# is needed in order to build the documentation
-import os
-import sys
+# import os
+# import sys
+import datetime
+import warnings
+
+warnings.filterwarnings(
+ "ignore",
+ category=UserWarning,
+ message="Matplotlib is currently using agg, which is a non-GUI backend, so cannot show the figure.",
+)
# local module should not be added to sys path if it's installed on RTFD
# see: https://stackoverflow.com/a/31882049/6696397
@@ -59,6 +67,7 @@ def setup(app):
"sphinx.ext.autosummary",
"sphinx.ext.napoleon", # parameters look better than with numpydoc only
"numpydoc",
+ "sphinx_gallery.gen_gallery",
]
# autosummaries from source-files
@@ -95,8 +104,9 @@ def setup(app):
master_doc = "contents"
# General information about the project.
+curr_year = datetime.datetime.now().year
project = "GSTools"
-copyright = "2018 - 2019, Lennart Schueler, Sebastian Mueller"
+copyright = "2018 - {}, Lennart Schueler, Sebastian Mueller".format(curr_year)
author = "Lennart Schueler, Sebastian Mueller"
# The version info for the project you're documenting, acts as replacement for
@@ -171,7 +181,9 @@ def setup(app):
# Output file base name for HTML help builder.
htmlhelp_basename = "GeoStatToolsdoc"
-
+# logos for the page
+html_logo = "pics/gstools_150.png"
+html_favicon = "pics/gstools.ico"
# -- Options for LaTeX output ---------------------------------------------
# latex_engine = 'lualatex'
@@ -247,3 +259,48 @@ def setup(app):
"hankel": ("https://hankel.readthedocs.io/en/latest/", None),
"emcee": ("https://emcee.readthedocs.io/en/latest/", None),
}
+
+
+# -- Sphinx Gallery Options
+from sphinx_gallery.sorting import FileNameSortKey
+
+sphinx_gallery_conf = {
+ # only show "print" output as output
+ "capture_repr": (),
+ # path to your examples scripts
+ "examples_dirs": [
+ "../../examples/00_misc/",
+ "../../examples/01_random_field/",
+ "../../examples/02_cov_model/",
+ "../../examples/03_variogram/",
+ "../../examples/04_vector_field/",
+ "../../examples/05_kriging/",
+ "../../examples/06_conditioned_fields/",
+ "../../examples/07_transformations/",
+ ],
+ # path where to save gallery generated examples
+ "gallery_dirs": [
+ "examples/00_misc/",
+ "examples/01_random_field/",
+ "examples/02_cov_model/",
+ "examples/03_variogram/",
+ "examples/04_vector_field/",
+ "examples/05_kriging/",
+ "examples/06_conditioned_fields/",
+ "examples/07_transformations/",
+ ],
+ # Pattern to search for example files
+ "filename_pattern": r"\.py",
+ # Remove the "Download all examples" button from the top level gallery
+ "download_all_examples": False,
+ # Sort gallery example by file name instead of number of lines (default)
+ "within_subsection_order": FileNameSortKey,
+ # directory where function granular galleries are stored
+ "backreferences_dir": None,
+ # Modules for which function level galleries are created. In
+ "doc_module": "gstools",
+ # "image_scrapers": ('pyvista', 'matplotlib'),
+ # "first_notebook_cell": ("%matplotlib inline\n"
+ # "from pyvista import set_plot_theme\n"
+ # "set_plot_theme('document')"),
+}
diff --git a/docs/source/covmodel.models.rst b/docs/source/covmodel.models.rst
deleted file mode 100644
index faffb056..00000000
--- a/docs/source/covmodel.models.rst
+++ /dev/null
@@ -1,12 +0,0 @@
-gstools.covmodel.models
------------------------
-
-.. automodule:: gstools.covmodel.models
- :members:
- :undoc-members:
- :no-inherited-members:
- :show-inheritance:
-
-.. raw:: latex
-
- \clearpage
diff --git a/docs/source/covmodel.rst b/docs/source/covmodel.rst
index 37b22dbd..74e7cebe 100644
--- a/docs/source/covmodel.rst
+++ b/docs/source/covmodel.rst
@@ -10,7 +10,4 @@ gstools.covmodel
.. toctree::
:hidden:
- covmodel.base.rst
- covmodel.models.rst
- covmodel.tpl_models.rst
covmodel.plot.rst
diff --git a/docs/source/covmodel.tpl_models.rst b/docs/source/covmodel.tpl_models.rst
deleted file mode 100644
index dd57c096..00000000
--- a/docs/source/covmodel.tpl_models.rst
+++ /dev/null
@@ -1,11 +0,0 @@
-gstools.covmodel.tpl_models
----------------------------
-
-.. automodule:: gstools.covmodel.tpl_models
- :members:
- :undoc-members:
- :show-inheritance:
-
-.. raw:: latex
-
- \clearpage
diff --git a/docs/source/index.rst b/docs/source/index.rst
index 67dd85c4..bc3d23e9 100644
--- a/docs/source/index.rst
+++ b/docs/source/index.rst
@@ -48,24 +48,28 @@ To get the latest development version you can install it directly from GitHub:
.. code-block:: none
- pip install https://github.com/GeoStat-Framework/GSTools/archive/develop.zip
+ pip install git+git://github.com/GeoStat-Framework/GSTools.git@develop
+
+If something went wrong during installation, try the :code:`-I` `flag from pip `_.
To enable the OpenMP support, you have to provide a C compiler, Cython and OpenMP.
To get all other dependencies, it is recommended to first install gstools once
in the standard way just decribed.
-Then use the following command:
+Simply use the following commands:
.. code-block:: none
- pip install --global-option="--openmp" gstools
+ pip install gstools
+ pip install -I --no-deps --global-option="--openmp" gstools
Or for the development version:
.. code-block:: none
- pip install --global-option="--openmp" https://github.com/GeoStat-Framework/GSTools/archive/develop.zip
+ pip install git+git://github.com/GeoStat-Framework/GSTools.git@develop
+ pip install -I --no-deps --global-option="--openmp" git+git://github.com/GeoStat-Framework/GSTools.git@develop
-If something went wrong during installation, try the :code:`-I` `flag from pip `_.
+The flags :code:`-I --no-deps` force pip to reinstall gstools but not the dependencies.
Citation
@@ -73,23 +77,27 @@ Citation
At the moment you can cite the Zenodo code publication of GSTools:
-| *Sebastian Müller, & Lennart Schüler. (2019, October 1). GeoStat-Framework/GSTools: Reverberating Red (Version v1.1.0). Zenodo. http://doi.org/10.5281/zenodo.3468230*
+| *Sebastian Müller & Lennart Schüler. GeoStat-Framework/GSTools. Zenodo. https://doi.org/10.5281/zenodo.1313628*
+
+If you want to cite a specific version, have a look at the Zenodo site.
A publication for the GeoStat-Framework is in preperation.
+
Tutorials and Examples
======================
-The documentation also includes some `tutorials `_,
+The documentation also includes some `tutorials `__,
showing the most important use cases of GSTools, which are
-- `Random Field Generation `_
-- `The Covariance Model `_
-- `Variogram Estimation `_
-- `Random Vector Field Generation `_
-- `Kriging `_
-- `Conditioned random field generation `_
-- `Field transformations `_
+- `Random Field Generation `__
+- `The Covariance Model `__
+- `Variogram Estimation `__
+- `Random Vector Field Generation `__
+- `Kriging `__
+- `Conditioned random field generation `__
+- `Field transformations `__
+- `Miscellaneous examples `__
Some more examples are provided in the examples folder.
@@ -105,7 +113,6 @@ These fields are generated using the randomisation method, described by
Examples
--------
-
Gaussian Covariance Model
^^^^^^^^^^^^^^^^^^^^^^^^^
@@ -114,12 +121,11 @@ with a :any:`Gaussian` covariance model.
.. code-block:: python
- from gstools import SRF, Gaussian
- import matplotlib.pyplot as plt
+ import gstools as gs
# structured field with a size 100x100 and a grid-size of 1x1
x = y = range(100)
- model = Gaussian(dim=2, var=1, len_scale=10)
- srf = SRF(model)
+ model = gs.Gaussian(dim=2, var=1, len_scale=10)
+ srf = gs.SRF(model)
srf((x, y), mesh_type='structured')
srf.plot()
@@ -134,12 +140,11 @@ A similar example but for a three dimensional field is exported to a
.. code-block:: python
- from gstools import SRF, Gaussian
- import matplotlib.pyplot as pt
+ import gstools as gs
# structured field with a size 100x100x100 and a grid-size of 1x1x1
x = y = z = range(100)
- model = Gaussian(dim=3, var=0.6, len_scale=20)
- srf = SRF(model)
+ model = gs.Gaussian(dim=3, var=0.6, len_scale=20)
+ srf = gs.SRF(model)
srf((x, y, z), mesh_type='structured')
srf.vtk_export('3d_field') # Save to a VTK file for ParaView
@@ -151,70 +156,6 @@ A similar example but for a three dimensional field is exported to a
:align: center
-Truncated Power Law Model
-^^^^^^^^^^^^^^^^^^^^^^^^^
-
-GSTools also implements truncated power law variograms, which can be represented as a
-superposition of scale dependant modes in form of standard variograms, which are truncated by
-a lower- :math:`\ell_{\mathrm{low}}` and an upper length-scale :math:`\ell_{\mathrm{up}}`.
-
-This example shows the truncated power law (:any:`TPLStable`) based on the
-:any:`Stable` covariance model and is given by
-
-.. math::
- \gamma_{\ell_{\mathrm{low}},\ell_{\mathrm{up}}}(r) =
- \intop_{\ell_{\mathrm{low}}}^{\ell_{\mathrm{up}}}
- \gamma(r,\lambda) \frac{\rm d \lambda}{\lambda}
-
-with `Stable` modes on each scale:
-
-.. math::
- \gamma(r,\lambda) &=
- \sigma^2(\lambda)\cdot\left(1-
- \exp\left[- \left(\frac{r}{\lambda}\right)^{\alpha}\right]
- \right)\\
- \sigma^2(\lambda) &= C\cdot\lambda^{2H}
-
-which gives Gaussian modes for ``alpha=2`` or Exponential modes for ``alpha=1``.
-
-For :math:`\ell_{\mathrm{low}}=0` this results in:
-
-.. math::
- \gamma_{\ell_{\mathrm{up}}}(r) &=
- \sigma^2_{\ell_{\mathrm{up}}}\cdot\left(1-
- \frac{2H}{\alpha} \cdot
- E_{1+\frac{2H}{\alpha}}
- \left[\left(\frac{r}{\ell_{\mathrm{up}}}\right)^{\alpha}\right]
- \right) \\
- \sigma^2_{\ell_{\mathrm{up}}} &=
- C\cdot\frac{\ell_{\mathrm{up}}^{2H}}{2H}
-
-.. code-block:: python
-
- import numpy as np
- import matplotlib.pyplot as plt
- from gstools import SRF, TPLStable
- x = y = np.linspace(0, 100, 100)
- model = TPLStable(
- dim=2, # spatial dimension
- var=1, # variance (C calculated internally, so that `var` is 1)
- len_low=0, # lower truncation of the power law
- len_scale=10, # length scale (a.k.a. range), len_up = len_low + len_scale
- nugget=0.1, # nugget
- anis=0.5, # anisotropy between main direction and transversal ones
- angles=np.pi/4, # rotation angles
- alpha=1.5, # shape parameter from the stable model
- hurst=0.7, # hurst coefficient from the power law
- )
- srf = SRF(model, mean=1, mode_no=1000, seed=19970221, verbose=True)
- srf((x, y), mesh_type='structured')
- srf.plot()
-
-.. image:: https://raw.githubusercontent.com/GeoStat-Framework/GSTools/master/docs/source/pics/tplstable_field.png
- :width: 400px
- :align: center
-
-
Estimating and fitting variograms
=================================
@@ -232,18 +173,18 @@ model again.
.. code-block:: python
import numpy as np
- from gstools import SRF, Exponential, Stable, vario_estimate_unstructured
+ import gstools as gs
# generate a synthetic field with an exponential model
x = np.random.RandomState(19970221).rand(1000) * 100.
y = np.random.RandomState(20011012).rand(1000) * 100.
- model = Exponential(dim=2, var=2, len_scale=8)
- srf = SRF(model, mean=0, seed=19970221)
+ model = gs.Exponential(dim=2, var=2, len_scale=8)
+ srf = gs.SRF(model, mean=0, seed=19970221)
field = srf((x, y))
# estimate the variogram of the field with 40 bins
bins = np.arange(40)
- bin_center, gamma = vario_estimate_unstructured((x, y), field, bins)
+ bin_center, gamma = gs.vario_estimate_unstructured((x, y), field, bins)
# fit the variogram with a stable model. (no nugget fitted)
- fit_model = Stable(dim=2)
+ fit_model = gs.Stable(dim=2)
fit_model.fit_variogram(bin_center, gamma, nugget=False)
# output
ax = fit_model.plot(x_max=40)
@@ -268,6 +209,7 @@ An important part of geostatistics is Kriging and conditioning spatial random
fields to measurements. With conditioned random fields, an ensemble of field realizations
with their variability depending on the proximity of the measurements can be generated.
+
Example
-------
@@ -277,8 +219,8 @@ generate 100 realizations and plot them:
.. code-block:: python
import numpy as np
- from gstools import Gaussian, SRF
import matplotlib.pyplot as plt
+ import gstools as gs
# conditions
cond_pos = [0.3, 1.9, 1.1, 3.3, 4.7]
@@ -287,8 +229,8 @@ generate 100 realizations and plot them:
gridx = np.linspace(0.0, 15.0, 151)
# spatial random field class
- model = Gaussian(dim=1, var=0.5, len_scale=2)
- srf = SRF(model)
+ model = gs.Gaussian(dim=1, var=0.5, len_scale=2)
+ srf = gs.SRF(model)
srf.set_condition(cond_pos, cond_val, "ordinary")
# generate the ensemble of field realizations
@@ -321,10 +263,10 @@ which takes a non-dimensional distance :class:`h = r/l`
.. code-block:: python
- from gstools import CovModel
import numpy as np
+ import gstools as gs
# use CovModel as the base-class
- class Gau(CovModel):
+ class Gau(gs.CovModel):
def cor(self, h):
return np.exp(-h**2)
@@ -345,12 +287,11 @@ Example
.. code-block:: python
import numpy as np
- import matplotlib.pyplot as plt
- from gstools import SRF, Gaussian
+ import gstools as gs
x = np.arange(100)
y = np.arange(100)
- model = Gaussian(dim=2, var=1, len_scale=10)
- srf = SRF(model, generator='VectorField')
+ model = gs.Gaussian(dim=2, var=1, len_scale=10)
+ srf = gs.SRF(model, generator='VectorField')
srf((x, y), mesh_type='structured', seed=19841203)
srf.plot()
@@ -370,10 +311,10 @@ create a VTK/PyVista dataset for use in Python with to :class:`.to_pyvista()` me
.. code-block:: python
- from gstools import SRF, Gaussian
+ import gstools as gs
x = y = range(100)
- model = Gaussian(dim=2, var=1, len_scale=10)
- srf = SRF(model)
+ model = gs.Gaussian(dim=2, var=1, len_scale=10)
+ srf = gs.SRF(model)
srf((x, y), mesh_type='structured')
srf.vtk_export("field") # Saves to a VTK file
mesh = srf.to_pyvista() # Create a VTK/PyVista dataset in memory
@@ -382,16 +323,20 @@ create a VTK/PyVista dataset for use in Python with to :class:`.to_pyvista()` me
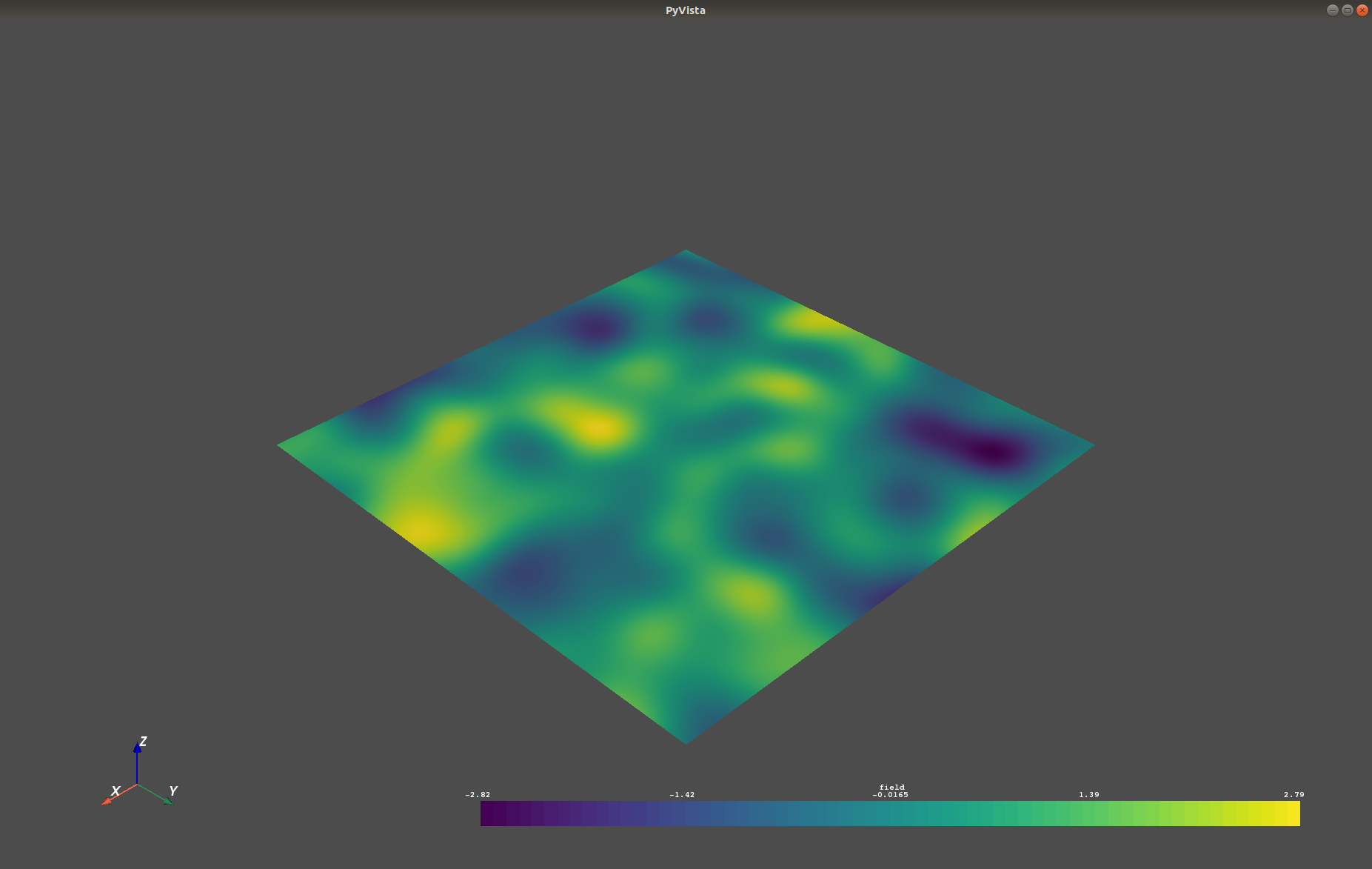
Which gives a RectilinearGrid VTK file :file:`field.vtr` or creates a PyVista mesh
in memory for immediate 3D plotting in Python.
+.. image:: https://raw.githubusercontent.com/GeoStat-Framework/GSTools/master/docs/source/pics/pyvista_export.png
+ :width: 600px
+ :align: center
+
Requirements
============
- `Numpy >= 1.14.5 `_
- `SciPy >= 1.1.0 `_
-- `hankel >= 0.3.6 `_
+- `hankel >= 1.0.2 `_
- `emcee >= 3.0.0 `_
-- `pyevtk `_
-- `six `_
+- `pyevtk >= 1.1.1 `_
+
Optional
--------
@@ -403,4 +348,4 @@ Optional
License
=======
-`LGPLv3 `_ © 2018-2019
+`LGPLv3 `_
diff --git a/docs/source/krige.rst b/docs/source/krige.rst
index 148c5c8d..e7eb6bd4 100644
--- a/docs/source/krige.rst
+++ b/docs/source/krige.rst
@@ -2,10 +2,6 @@ gstools.krige
=============
.. automodule:: gstools.krige
- :members:
- :undoc-members:
- :inherited-members:
- :show-inheritance:
.. raw:: latex
diff --git a/docs/source/pics/gstools.ico b/docs/source/pics/gstools.ico
new file mode 100644
index 00000000..9119caaa
Binary files /dev/null and b/docs/source/pics/gstools.ico differ
diff --git a/docs/source/tutorial_01_srf.rst b/docs/source/tutorial_01_srf.rst
deleted file mode 100644
index 9ce2164e..00000000
--- a/docs/source/tutorial_01_srf.rst
+++ /dev/null
@@ -1,281 +0,0 @@
-Tutorial 1: Random Field Generation
-===================================
-
-The main feature of GSTools is the spatial random field generator :any:`SRF`,
-which can generate random fields following a given covariance model.
-The generator provides a lot of nice features, which will be explained in
-the following
-
-Theoretical Background
-----------------------
-
-GSTools generates spatial random fields with a given covariance model or
-semi-variogram. This is done by using the so-called randomization method.
-The spatial random field is represented by a stochastic Fourier integral
-and its discretised modes are evaluated at random frequencies.
-
-GSTools supports arbitrary and non-isotropic covariance models.
-
-A very Simple Example
----------------------
-
-We are going to start with a very simple example of a spatial random field
-with an isotropic Gaussian covariance model and following parameters:
-
-- variance :math:`\sigma^2=1`
-- correlation length :math:`\lambda=10`
-
-First, we set things up and create the axes for the field. We are going to
-need the :any:`SRF` class for the actual generation of the spatial random field.
-But :any:`SRF` also needs a covariance model and we will simply take the :any:`Gaussian` model.
-
-.. code-block:: python
-
- from gstools import SRF, Gaussian
-
- x = y = range(100)
-
-Now we create the covariance model with the parameters :math:`\sigma^2` and
-:math:`\lambda` and hand it over to :any:`SRF`. By specifying a seed,
-we make sure to create reproducible results:
-
-.. code-block:: python
-
- model = Gaussian(dim=2, var=1, len_scale=10)
- srf = SRF(model, seed=20170519)
-
-With these simple steps, everything is ready to create our first random field.
-We will create the field on a structured grid (as you might have guessed from the `x` and `y`), which makes it easier to plot.
-
-.. code-block:: python
-
- field = srf.structured([x, y])
- srf.plot()
-
-Yielding
-
-.. image:: pics/srf_tut_gau_field.png
- :width: 600px
- :align: center
-
-Wow, that was pretty easy!
-
-The script can be found in :download:`gstools/examples/00_gaussian.py<../../examples/00_gaussian.py>`
-
-Creating an Ensemble of Fields
-------------------------------
-
-Creating an ensemble of random fields would also be
-a great idea. Let's reuse most of the previous code.
-
-.. code-block:: python
-
- import numpy as np
- import matplotlib.pyplot as pt
- from gstools import SRF, Gaussian
-
- x = y = np.arange(100)
-
- model = Gaussian(dim=2, var=1, len_scale=10)
- srf = SRF(model)
-
-This time, we did not provide a seed to :any:`SRF`, as the seeds will used
-during the actual computation of the fields. We will create four ensemble
-members, for better visualisation and save them in a list and in a first
-step, we will be using the loop counter as the seeds.
-
-.. code-block:: python
-
- ens_no = 4
- field = []
- for i in range(ens_no):
- field.append(srf.structured([x, y], seed=i))
-
-Now let's have a look at the results:
-
-.. code-block:: python
-
- fig, ax = pt.subplots(2, 2, sharex=True, sharey=True)
- ax = ax.flatten()
- for i in range(ens_no):
- ax[i].imshow(field[i].T, origin='lower')
- pt.show()
-
-Yielding
-
-.. image:: pics/srf_tut_gau_field_ens.png
- :width: 600px
- :align: center
-
-The script can be found in :download:`gstools/examples/05_srf_ensemble.py<../../examples/05_srf_ensemble.py>`
-
-Using better Seeds
-^^^^^^^^^^^^^^^^^^
-
-It is not always a good idea to use incrementing seeds. Therefore GSTools
-provides a seed generator :any:`MasterRNG`. The loop, in which the fields are generated would
-then look like
-
-.. code-block:: python
-
- from gstools.random import MasterRNG
- seed = MasterRNG(20170519)
- for i in range(ens_no):
- field.append(srf.structured([x, y], seed=seed()))
-
-Creating Fancier Fields
------------------------
-
-Only using Gaussian covariance fields gets boring. Now we are going to create much rougher random fields by using an exponential covariance model and we are going to make them anisotropic.
-
-The code is very similar to the previous examples, but with a different covariance model class :any:`Exponential`. As model parameters we a using following
-
-- variance :math:`\sigma^2=1`
-- correlation length :math:`\lambda=(12, 3)^T`
-- rotation angle :math:`\theta=\pi/8`
-
-
-.. code-block:: python
-
- import numpy as np
- from gstools import SRF, Exponential
-
- x = y = np.arange(100)
-
- model = Exponential(dim=2, var=1, len_scale=[12., 3.], angles=np.pi/8.)
- srf = SRF(model, seed=20170519)
-
- srf.structured([x, y])
- srf.plot()
-
-Yielding
-
-.. image:: pics/srf_tut_exp_ani_rot.png
- :width: 600px
- :align: center
-
-The anisotropy ratio could also have been set with
-
-.. code-block:: python
-
- model = Exponential(dim=2, var=1, len_scale=12., anis=3./12., angles=np.pi/8.)
-
-Using an Unstructured Grid
---------------------------
-
-For many applications, the random fields are needed on an unstructured grid.
-Normally, such a grid would be read in, but we can simply generate one and
-then create a random field at those coordinates.
-
-.. code-block:: python
-
- import numpy as np
- from gstools import SRF, Exponential
- from gstools.random import MasterRNG
-
- seed = MasterRNG(19970221)
- rng = np.random.RandomState(seed())
- x = rng.randint(0, 100, size=10000)
- y = rng.randint(0, 100, size=10000)
-
- model = Exponential(dim=2, var=1, len_scale=[12., 3.], angles=np.pi/8.)
-
- srf = SRF(model, seed=20170519)
- srf([x, y])
- srf.plot()
-
-Yielding
-
-.. image:: pics/srf_tut_unstr.png
- :width: 600px
- :align: center
-
-Comparing this image to the previous one, you can see that be using the same
-seed, the same field can be computed on different grids.
-
-The script can be found in :download:`gstools/examples/06_unstr_srf_export.py<../../examples/06_unstr_srf_export.py>`
-
-Exporting a Field
------------------
-
-Using the field from `previous example `__, it can simply be exported to the file
-``field.vtu`` and viewed by e.g. paraview with following lines of code
-
-.. code-block:: python
-
- srf.vtk_export("field")
-
-Or it could visualized immediately in Python using `PyVista `__:
-
-.. code-block:: python
-
- mesh = srf.to_pyvista("field")
- mesh.plot()
-
-The script can be found in :download:`gstools/examples/04_export.py<../../examples/04_export.py>` and
-in :download:`gstools/examples/06_unstr_srf_export.py<../../examples/06_unstr_srf_export.py>`
-
-Merging two Fields
-------------------
-
-We can even generate the same field realisation on different grids. Let's try
-to merge two unstructured rectangular fields. The first field will be generated
-exactly like in example `Using an Unstructured Grid`_:
-
-.. code-block:: python
-
- import numpy as np
- import matplotlib.pyplot as pt
- from gstools import SRF, Exponential
- from gstools.random import MasterRNG
-
- seed = MasterRNG(19970221)
- rng = np.random.RandomState(seed())
- x = rng.randint(0, 100, size=10000)
- y = rng.randint(0, 100, size=10000)
-
- model = Exponential(dim=2, var=1, len_scale=[12., 3.], angles=np.pi/8.)
-
- srf = SRF(model, seed=20170519)
-
- field = srf([x, y])
-
-But now we extend the field on the right hand side by creating a new
-unstructured grid and calculating a field with the same parameters and the
-same seed on it:
-
-.. code-block:: python
-
- # new grid
- seed = MasterRNG(20011012)
- rng = np.random.RandomState(seed())
- x2 = rng.randint(99, 150, size=10000)
- y2 = rng.randint(20, 80, size=10000)
-
- field2 = srf((x2, y2))
-
- pt.tricontourf(x, y, field.T)
- pt.tricontourf(x2, y2, field2.T)
- pt.axes().set_aspect('equal')
- pt.show()
-
-Yielding
-
-.. image:: pics/srf_tut_merge.png
- :width: 600px
- :align: center
-
-The slight mismatch where the two fields were merged is merely due to
-interpolation problems of the plotting routine. You can convince yourself
-be increasing the resolution of the grids by a factor of 10.
-
-Of course, this merging could also have been done by appending the grid
-point ``(x2, y2)`` to the original grid ``(x, y)`` before generating the field.
-But one application scenario would be to generate hugh fields, which would not
-fit into memory anymore.
-
-The script can be found in :download:`gstools/examples/07_srf_merge.py<../../examples/07_srf_merge.py>`
-
-.. raw:: latex
-
- \clearpage
diff --git a/docs/source/tutorial_02_cov.rst b/docs/source/tutorial_02_cov.rst
deleted file mode 100644
index 0b1bd8d6..00000000
--- a/docs/source/tutorial_02_cov.rst
+++ /dev/null
@@ -1,500 +0,0 @@
-Tutorial 2: The Covariance Model
-================================
-
-One of the core-features of GSTools is the powerful :any:`CovModel`
-class, which allows you to easily define arbitrary covariance models by
-yourself. The resulting models provide a bunch of nice features to explore the
-covariance models.
-
-Theoretical Backgound
----------------------
-
-A covariance model is used to characterize the
-`semi-variogram `_,
-denoted by :math:`\gamma`, of a spatial random field.
-In GSTools, we use the following form for an isotropic and stationary field:
-
-.. math::
- \gamma\left(r\right)=
- \sigma^2\cdot\left(1-\mathrm{cor}\left(r\right)\right)+n
-
-Where:
- - :math:`\mathrm{cor}(r)` is the so called
- `correlation `_
- function depending on the distance :math:`r`
- - :math:`\sigma^2` is the variance
- - :math:`n` is the nugget (subscale variance)
-
-.. note::
-
- We are not limited to isotropic models. We support anisotropy ratios for
- length scales in orthogonal transversal directions like:
-
- - :math:`x` (main direction)
- - :math:`y` (1. transversal direction)
- - :math:`z` (2. transversal direction)
-
- These main directions can also be rotated, but we will come to that later.
-
-Example
--------
-
-Let us start with a short example of a self defined model (Of course, we
-provide a lot of predefined models [See: :any:`gstools.covmodel`],
-but they all work the same way).
-Therefore we reimplement the Gaussian covariance model by defining just the
-`correlation `_ function:
-
-.. code-block:: python
-
- from gstools import CovModel
- import numpy as np
- # use CovModel as the base-class
- class Gau(CovModel):
- def correlation(self, r):
- return np.exp(-(r/self.len_scale)**2)
-
-Now we can instantiate this model:
-
-.. code-block:: python
-
- model = Gau(dim=2, var=2., len_scale=10)
-
-To have a look at the variogram, let's plot it:
-
-.. code-block:: python
-
- from gstools.covmodel.plot import plot_variogram
- plot_variogram(model)
-
-Which gives:
-
-.. image:: pics/cov_model_vario.png
- :width: 400px
- :align: center
-
-Parameters
-----------
-
-We already used some parameters, which every covariance models has. The basic ones
-are:
-
- - **dim** : dimension of the model
- - **var** : variance of the model (on top of the subscale variance)
- - **len_scale** : length scale of the model
- - **nugget** : nugget (subscale variance) of the model
-
-These are the common parameters used to characterize a covariance model and are
-therefore used by every model in GSTools. You can also access and reset them:
-
-.. code-block:: python
-
- print(model.dim, model.var, model.len_scale, model.nugget, model.sill)
- model.dim = 3
- model.var = 1
- model.len_scale = 15
- model.nugget = 0.1
- print(model.dim, model.var, model.len_scale, model.nugget, model.sill)
-
-Which gives:
-
-.. code-block:: python
-
- 2 2.0 10 0.0 2.0
- 3 1.0 15 0.1 1.1
-
-.. note::
-
- - The sill of the variogram is calculated by ``sill = variance + nugget``
- So we treat the variance as everything **above** the nugget, which is sometimes
- called **partial sill**.
- - A covariance model can also have additional parameters.
-
-Anisotropy
-----------
-
-The internally used (semi-) variogram represents the isotropic case for the model.
-Nevertheless, you can provide anisotropy ratios by:
-
-.. code-block:: python
-
- model = Gau(dim=3, var=2., len_scale=10, anis=0.5)
- print(model.anis)
- print(model.len_scale_vec)
-
-Which gives:
-
-.. code-block:: python
-
- [0.5 1. ]
- [10. 5. 10.]
-
-As you can see, we defined just one anisotropy-ratio and the second transversal
-direction was filled up with ``1.`` and you can get the length-scales in each
-direction by the attribute :any:`len_scale_vec`. For full control you can set
-a list of anistropy ratios: ``anis=[0.5, 0.4]``.
-
-Alternatively you can provide a list of length-scales:
-
-.. code-block:: python
-
- model = Gau(dim=3, var=2., len_scale=[10, 5, 4])
- print(model.anis)
- print(model.len_scale)
- print(model.len_scale_vec)
-
-Which gives:
-
-.. code-block:: python
-
- [0.5 0.4]
- 10
- [10. 5. 4.]
-
-Rotation Angles
----------------
-
-The main directions of the field don't have to coincide with the spatial
-directions :math:`x`, :math:`y` and :math:`z`. Therefore you can provide
-rotation angles for the model:
-
-.. code-block:: python
-
- model = Gau(dim=3, var=2., len_scale=10, angles=2.5)
- print(model.angles)
-
-Which gives:
-
-.. code-block:: python
-
- [2.5 0. 0. ]
-
-Again, the angles were filled up with ``0.`` to match the dimension and you
-could also provide a list of angles. The number of angles depends on the
-given dimension:
-
-- in 1D: no rotation performable
-- in 2D: given as rotation around z-axis
-- in 3D: given by yaw, pitch, and roll (known as
- `Tait–Bryan `_
- angles)
-
-Methods
--------
-
-The covariance model class :any:`CovModel` of GSTools provides a set of handy
-methods.
-
-Basics
-^^^^^^
-
-One of the following functions defines the main characterization of the
-variogram:
-
-- ``variogram`` : The variogram of the model given by
-
- .. math::
- \gamma\left(r\right)=
- \sigma^2\cdot\left(1-\mathrm{cor}\left(r\right)\right)+n
-
-- ``covariance`` : The (auto-)covariance of the model given by
-
- .. math::
- C\left(r\right)= \sigma^2\cdot\mathrm{cor}\left(r\right)
-
-- ``correlation`` : The (auto-)correlation (or normalized covariance)
- of the model given by
-
- .. math::
- \mathrm{cor}\left(r\right)
-
-As you can see, it is the easiest way to define a covariance model by giving a
-correlation function as demonstrated by the above model ``Gau``.
-If one of the above functions is given, the others will be determined:
-
-.. code-block:: python
-
- model = Gau(dim=3, var=2., len_scale=10, nugget=0.5)
- print(model.variogram(10.))
- print(model.covariance(10.))
- print(model.correlation(10.))
-
-Which gives:
-
-.. code-block:: python
-
- 1.7642411176571153
- 0.6321205588285577
- 0.7357588823428847
- 0.36787944117144233
-
-Spectral methods
-^^^^^^^^^^^^^^^^
-
-The spectrum of a covariance model is given by:
-
-.. math:: S(\mathbf{k}) = \left(\frac{1}{2\pi}\right)^n
- \int C(\Vert\mathbf{r}\Vert) e^{i b\mathbf{k}\cdot\mathbf{r}} d^n\mathbf{r}
-
-Since the covariance function :math:`C(r)` is radially symmetric, we can
-calculate this by the
-`hankel-transformation `_:
-
-.. math:: S(k) = \left(\frac{1}{2\pi}\right)^n \cdot
- \frac{(2\pi)^{n/2}}{(bk)^{n/2-1}}
- \int_0^\infty r^{n/2-1} C(r) J_{n/2-1}(bkr) r dr
-
-Where :math:`k=\left\Vert\mathbf{k}\right\Vert`.
-
-Depending on the spectrum, the spectral-density is defined by:
-
-.. math:: \tilde{S}(k) = \frac{S(k)}{\sigma^2}
-
-You can access these methods by:
-
-.. code-block:: python
-
- model = Gau(dim=3, var=2., len_scale=10)
- print(model.spectrum(0.1))
- print(model.spectral_density(0.1))
-
-Which gives:
-
-.. code-block:: python
-
- 34.96564773852395
- 17.482823869261974
-
-.. note::
- The spectral-density is given by the radius of the input phase. But it is
- **not** a probability density function for the radius of the phase.
- To obtain the pdf for the phase-radius, you can use the methods
- :any:`spectral_rad_pdf` or :any:`ln_spectral_rad_pdf` for the logarithm.
-
- The user can also provide a cdf (cumulative distribution function) by
- defining a method called ``spectral_rad_cdf`` and/or a ppf (percent-point function)
- by ``spectral_rad_ppf``.
-
- The attributes :any:`has_cdf` and :any:`has_ppf` will check for that.
-
-Different scales
-----------------
-
-Besides the length-scale, there are many other ways of characterizing a certain
-scale of a covariance model. We provide two common scales with the covariance
-model.
-
-Integral scale
-^^^^^^^^^^^^^^
-
-The `integral scale `_
-of a covariance model is calculated by:
-
-.. math:: I = \int_0^\infty \mathrm{cor}(r) dr
-
-You can access it by:
-
-.. code-block:: python
-
- model = Gau(dim=3, var=2., len_scale=10)
- print(model.integral_scale)
- print(model.integral_scale_vec)
-
-Which gives:
-
-.. code-block:: python
-
- 8.862269254527579
- [8.86226925 8.86226925 8.86226925]
-
-You can also specify integral length scales like the ordinary length scale,
-and len_scale/anis will be recalculated:
-
-.. code-block:: python
-
- model = Gau(dim=3, var=2., integral_scale=[10, 4, 2])
- print(model.anis)
- print(model.len_scale)
- print(model.len_scale_vec)
- print(model.integral_scale)
- print(model.integral_scale_vec)
-
-Which gives:
-
-.. code-block:: python
-
- [0.4 0.2]
- 11.283791670955127
- [11.28379167 4.51351667 2.25675833]
- 10.000000000000002
- [10. 4. 2.]
-
-Percentile scale
-^^^^^^^^^^^^^^^^
-
-Another scale characterizing the covariance model, is the percentile scale.
-It is the distance, where the normalized variogram reaches a certain percentage
-of its sill.
-
-.. code-block:: python
-
- model = Gau(dim=3, var=2., len_scale=10)
- print(model.percentile_scale(0.9))
-
-Which gives:
-
-.. code-block:: python
-
- 15.174271293851463
-
-.. note::
-
- The nugget is neglected by this percentile_scale.
-
-Additional Parameters
----------------------
-
-Let's pimp our self-defined model ``Gau`` by setting the exponent as an additional
-parameter:
-
-.. math:: \mathrm{cor}(r) := \exp\left(-\left(\frac{r}{\ell}\right)^{\alpha}\right)
-
-This leads to the so called **stable** covariance model and we can define it by
-
-.. code-block:: python
-
- class Stab(CovModel):
- def default_opt_arg(self):
- return {"alpha": 1.5}
- def correlation(self, r):
- return np.exp(-(r/self.len_scale)**self.alpha)
-
-As you can see, we override the method :any:`CovModel.default_opt_arg` to provide
-a standard value for the optional argument ``alpha`` and we can access it
-in the correlation function by ``self.alpha``
-
-Now we can instantiate this model:
-
-.. code-block:: python
-
- model1 = Stab(dim=2, var=2., len_scale=10)
- model2 = Stab(dim=2, var=2., len_scale=10, alpha=0.5)
- print(model1)
- print(model2)
-
-Which gives:
-
-.. code-block:: python
-
- Stab(dim=2, var=2.0, len_scale=10, nugget=0.0, anis=[1.], angles=[0.], alpha=1.5)
- Stab(dim=2, var=2.0, len_scale=10, nugget=0.0, anis=[1.], angles=[0.], alpha=0.5)
-
-.. note::
-
- You don't have to overrid the :any:`CovModel.default_opt_arg`, but you will
- get a ValueError if you don't set it on creation.
-
-Fitting variogram data
-----------------------
-
-The model class comes with a routine to fit the model-parameters to given
-variogram data. Have a look at the following:
-
-.. code-block:: python
-
- # data
- x = [1.0, 3.0, 5.0, 7.0, 9.0, 11.0]
- y = [0.2, 0.5, 0.6, 0.8, 0.8, 0.9]
- # fitting model
- model = Stab(dim=2)
- # we have to provide boundaries for the parameters
- model.set_arg_bounds(alpha=[0, 3])
- # fit the model to given data, deselect nugget
- results, pcov = model.fit_variogram(x, y, nugget=False)
- print(results)
- # show the fitting
- from matplotlib import pyplot as plt
- from gstools.covmodel.plot import plot_variogram
- plt.scatter(x, y, color="k")
- plot_variogram(model)
- plt.show()
-
-Which gives:
-
-.. code-block:: python
-
- {'var': 1.024575782651677,
- 'len_scale': 5.081620691462197,
- 'nugget': 0.0,
- 'alpha': 0.906705123369987}
-
-.. image:: pics/stab_vario_fit.png
- :width: 400px
- :align: center
-
-As you can see, we have to provide boundaries for the parameters.
-As a default, the following bounds are set:
-
-- additional parameters: ``[-np.inf, np.inf]``
-- variance: ``[0.0, np.inf]``
-- len_scale: ``[0.0, np.inf]``
-- nugget: ``[0.0, np.inf]``
-
-Also, you can deselect parameters from fitting, so their predefined values
-will be kept. In our case, we fixed a ``nugget`` of ``0.0``, which was set
-by default. You can deselect any standard or optional argument of the covariance model.
-The second return value ``pcov`` is the estimated covariance of ``popt`` from
-the used scipy routine :any:`scipy.optimize.curve_fit`.
-
-You can use the following methods to manipulate the used bounds:
-
-.. currentmodule:: gstools.covmodel.base
-
-.. autosummary::
- CovModel.default_opt_arg_bounds
- CovModel.default_arg_bounds
- CovModel.set_arg_bounds
- CovModel.check_arg_bounds
-
-You can override the :any:`CovModel.default_opt_arg_bounds` to provide standard
-bounds for your additional parameters.
-
-To access the bounds you can use:
-
-.. autosummary::
- CovModel.var_bounds
- CovModel.len_scale_bounds
- CovModel.nugget_bounds
- CovModel.opt_arg_bounds
- CovModel.arg_bounds
-
-Provided Covariance Models
---------------------------
-
-The following standard covariance models are provided by GSTools
-
-.. currentmodule:: gstools.covmodel.models
-
-.. autosummary::
- Gaussian
- Exponential
- Matern
- Stable
- Rational
- Linear
- Circular
- Spherical
- Intersection
-
-As a special feature, we also provide truncated power law (TPL) covariance models
-
-.. currentmodule:: gstools.covmodel.tpl_models
-
-.. autosummary::
- TPLGaussian
- TPLExponential
- TPLStable
-
-.. raw:: latex
-
- \clearpage
diff --git a/docs/source/tutorial_03_vario.rst b/docs/source/tutorial_03_vario.rst
deleted file mode 100644
index 6343985d..00000000
--- a/docs/source/tutorial_03_vario.rst
+++ /dev/null
@@ -1,356 +0,0 @@
-Tutorial 3: Variogram Estimation
-================================
-
-Estimating the spatial correlations is an important part of geostatistics.
-These spatial correlations can be expressed by the variogram, which can be
-estimated with the subpackage :any:`gstools.variogram`. The variograms can be
-estimated on structured and unstructured grids.
-
-Theoretical Background
-----------------------
-
-The same `(semi-)variogram `_ as
-:doc:`the Covariance Model` is being used
-by this subpackage.
-
-An Example with Actual Data
----------------------------
-
-This example is going to be a bit more extensive and we are going to do some
-basic data preprocessing for the actual variogram estimation. But this example
-will be self-contained and all data gathering and processing will be done in
-this example script.
-
-The complete script can be found in :download:`gstools/examples/08_variogram_estimation.py<../../examples/08_variogram_estimation.py>`
-
-*This example will only work with Python 3.*
-
-The Data
-^^^^^^^^
-
-We are going to analyse the Herten aquifer, which is situated in Southern
-Germany. Multiple outcrop faces where surveyed and interpolated to a 3D
-dataset. In these publications, you can find more information about the data:
-
-| Bayer, Peter; Comunian, Alessandro; Höyng, Dominik; Mariethoz, Gregoire (2015): Physicochemical properties and 3D geostatistical simulations of the Herten and the Descalvado aquifer analogs. PANGAEA, https://doi.org/10.1594/PANGAEA.844167,
-| Supplement to: Bayer, P et al. (2015): Three-dimensional multi-facies realizations of sedimentary reservoir and aquifer analogs. Scientific Data, 2, 150033, https://doi.org/10.1038/sdata.2015.33
-|
-
-Retrieving the Data
-^^^^^^^^^^^^^^^^^^^
-
-To begin with, we need to download and extract the data. Therefore, we are
-going to use some built-in Python libraries. For simplicity, many values and
-strings will be hardcoded.
-
-.. code-block:: python
-
- import os
- import urllib.request
- import zipfile
- import numpy as np
- import matplotlib.pyplot as pt
-
- def download_herten():
- # download the data, warning: its about 250MB
- print('Downloading Herten data')
- data_filename = 'data.zip'
- data_url = 'http://store.pangaea.de/Publications/Bayer_et_al_2015/Herten-analog.zip'
- urllib.request.urlretrieve(data_url, 'data.zip')
-
- # extract the data
- with zipfile.ZipFile(data_filename, 'r') as zf:
- zf.extract(os.path.join('Herten-analog', 'sim-big_1000x1000x140',
- 'sim.vtk'))
-
-That was that. But we also need a script to convert the data into a format we
-can use. This script is also kindly provided by the authors. We can download
-this script in a very similar manner as the data:
-
-.. code-block:: python
-
- def download_scripts():
- # download a script for file conversion
- print('Downloading scripts')
- tools_filename = 'scripts.zip'
- tool_url = 'http://store.pangaea.de/Publications/Bayer_et_al_2015/tools.zip'
- urllib.request.urlretrieve(tool_url, tools_filename)
-
- # only extract the script we need
- with zipfile.ZipFile(tools_filename, 'r') as zf:
- zf.extract(os.path.join('tools', 'vtk2gslib.py'))
-
-These two functions can now be called:
-
-.. code-block:: python
-
- download_herten()
- download_scripts()
-
-
-Preprocessing the Data
-^^^^^^^^^^^^^^^^^^^^^^
-
-First of all, we have to convert the data with the script we just downloaded
-
-.. code-block:: python
-
- # import the downloaded conversion script
- from tools.vtk2gslib import vtk2numpy
-
- # load the Herten aquifer with the downloaded vtk2numpy routine
- print('Loading data')
- herten, grid = vtk2numpy(os.path.join('Herten-analog', 'sim-big_1000x1000x140', 'sim.vtk'))
-
-The data only contains facies, but from the supplementary data, we know the
-hydraulic conductivity values of each facies, which we will simply paste here
-and assign them to the correct facies
-
-.. code-block:: python
-
- # conductivity values per fazies from the supplementary data
- cond = np.array([2.50E-04, 2.30E-04, 6.10E-05, 2.60E-02, 1.30E-01,
- 9.50E-02, 4.30E-05, 6.00E-07, 2.30E-03, 1.40E-04,])
-
- # asign the conductivities to the facies
- herten_cond = cond[herten]
-
-Next, we are going to calculate the transmissivity, by integrating over the
-vertical axis
-
-.. code-block:: python
-
- # integrate over the vertical axis, calculate transmissivity
- herten_log_trans = np.log(np.sum(herten_cond, axis=2) * grid['dz'])
-
-The Herten data provides information about the grid, which was already used in
-the previous code block. From this information, we can create our own grid on
-which we can estimate the variogram. As a first step, we are going to estimate
-an isotropic variogram, meaning that we will take point pairs from all
-directions into account. An unstructured grid is a natural choice for this.
-Therefore, we are going to create an unstructured grid from the given,
-structured one. For this, we are going to write another small function
-
-.. code-block:: python
-
- def create_unstructured_grid(x_s, y_s):
- x_u, y_u = np.meshgrid(x_s, y_s)
- len_unstruct = len(x_s) * len(y_s)
- x_u = np.reshape(x_u, len_unstruct)
- y_u = np.reshape(y_u, len_unstruct)
- return x_u, y_u
-
- # create a structured grid on which the data is defined
- x_s = np.arange(grid['ox'], grid['nx']*grid['dx'], grid['dx'])
- y_s = np.arange(grid['oy'], grid['ny']*grid['dy'], grid['dy'])
-
- # create an unstructured grid for the variogram estimation
- x_u, y_u = create_unstructured_grid(x_s, y_s)
-
-Let's have a look at the transmissivity field of the Herten aquifer
-
-.. code-block:: python
-
- pt.imshow(herten_log_trans.T, origin='lower', aspect='equal')
- pt.show()
-
-.. image:: pics/vario_tut_herten.png
- :width: 600px
- :align: center
-
-
-Estimating the Variogram
-^^^^^^^^^^^^^^^^^^^^^^^^
-
-Finally, everything is ready for the variogram estimation. For the unstructured
-method, we have to define the bins on which the variogram will be estimated.
-Through expert knowledge (i.e. fiddling around), we assume that the main
-features of the variogram will be below 10 metres distance. And because the
-data has a high spatial resolution, the resolution of the bins can also be
-high. The transmissivity data is still defined on a structured grid, but we can
-simply flatten it with :any:`numpy.ndarray.flatten`, in order to bring it into
-the right shape. It might be more memory efficient to use
-``herten_log_trans.reshape(-1)``, but for better readability, we will stick to
-:any:`numpy.ndarray.flatten`. Taking all data points into account would take a
-very long time (expert knowledge \*wink\*), thus we will only take 2000 datapoints into account, which are sampled randomly. In order to make the exact
-results reproducible, we can also set a seed.
-
-.. code-block:: python
-
- from gstools import vario_estimate_unstructured
-
- bins = np.linspace(0, 10, 50)
- print('Estimating unstructured variogram')
- bin_center, gamma = vario_estimate_unstructured(
- (x_u, y_u),
- herten_log_trans.flatten(),
- bins,
- sampling_size=2000,
- sampling_seed=19920516,
- )
-
-The estimated variogram is calculated on the centre of the given bins,
-therefore, the ``bin_center`` array is also returned.
-
-Fitting the Variogram
-^^^^^^^^^^^^^^^^^^^^^
-
-Now, we can see, if the estimated variogram can be modelled by a common
-variogram model. Let's try the :any:`Exponential` model.
-
-.. code-block:: python
-
- from gstools import Exponential
-
- # fit an exponential model
- fit_model = Exponential(dim=2)
- fit_model.fit_variogram(bin_center, gamma, nugget=False)
-
-Finally, we can visualise some results. For quickly plotting a covariance
-model, GSTools provides some helper functions.
-
-.. code-block:: python
-
- from gstools.covmodel.plot import plot_variogram
- pt.plot(bin_center, gamma)
- plot_variogram(fit_model, x_max=bins[-1])
- pt.show()
-
-.. image:: pics/vario_tut_fit_exp.png
- :width: 400px
- :align: center
-
-That looks like a pretty good fit! By printing the model, we can directly see
-the fitted parameters
-
-.. code-block:: python
-
- print(fit_model)
-
-which gives
-
-.. code-block:: python
-
- Exponential(dim=2, var=0.020193095802479327, len_scale=1.4480057557321007, nugget=0.0, anis=[1.], angles=[0.])
-
-With this data, we could start generating new ensembles of the Herten aquifer
-with the :any:`SRF` class.
-
-Estimating the Variogram in Specific Directions
-^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
-
-Estimating a variogram on a structured grid gives us the possibility to only
-consider values in a specific direction. This could be a first test, to see if
-the data is anisotropic.
-In order to speed up the calculations, we are going to only use every 10th datapoint and for a comparison with the isotropic variogram calculated earlier, we
-only need the first 21 array items.
-
-.. code-block:: python
-
- x_s = x_s[::10][:21]
- y_s = y_s[::10][:21]
- herten_trans_log = herten_log_trans[::10,::10]
-
-With this much smaller data set, we can immediately estimate the variogram in
-the x- and y-axis
-
-.. code-block:: python
-
- from gstools import vario_estimate_structured
- print('Estimating structured variograms')
- gamma_x = vario_estimate_structured(herten_trans_log, direction='x')[:21]
- gamma_y = vario_estimate_structured(herten_trans_log, direction='y')[:21]
-
-With these two estimated variograms, we can start fitting :any:`Exponential`
-covariance models
-
-.. code-block:: python
-
- fit_model_x = Exponential(dim=2)
- fit_model_x.fit_variogram(x_s, gamma_x, nugget=False)
- fit_model_y = Exponential(dim=2)
- fit_model_y.fit_variogram(y_s, gamma_y, nugget=False)
-
-Now, the isotropic variogram and the two variograms in x- and y-direction can
-be plotted together with their respective models, which will be plotted with
-dashed lines.
-
-.. code-block:: python
-
- line, = pt.plot(bin_center, gamma, label='estimated variogram (isotropic)')
- pt.plot(bin_center, fit_model.variogram(bin_center), color=line.get_color(),
- linestyle='--', label='exp. variogram (isotropic)')
-
- line, = pt.plot(x_s, gamma_x, label='estimated variogram in x-dir')
- pt.plot(x_s, fit_model_x.variogram(x_s), color=line.get_color(),
- linestyle='--', label='exp. variogram in x-dir')
-
- line, = pt.plot(y_s, gamma_y, label='estimated variogram in y-dir')
- pt.plot(y_s, fit_model_y.variogram(y_s),
- color=line.get_color(), linestyle='--', label='exp. variogram in y-dir')
-
- pt.legend()
- pt.show()
-
-Giving
-
-.. image:: pics/vario_tut_aniso_fit_exp.png
- :width: 400px
- :align: center
-
-The plot might be a bit cluttered, but at least it is pretty obvious that the
-Herten aquifer has no apparent anisotropies in its spatial structure.
-
-Creating a Spatial Random Field from the Herten Parameters
-^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
-
-With all the hard work done, it's straight forward now, to generate new
-*Herten realisations*
-
-.. code-block:: python
-
- from gstools import SRF
-
- srf = SRF(fit_model, seed=19770928)
- new_herten = srf((x_s, y_s), mesh_type='structured')
-
- pt.imshow(new_herten.T, origin='lower')
- pt.show()
-
-Yielding
-
-.. image:: pics/vario_tut_new_herten.png
- :width: 600px
- :align: center
-
-
-That's pretty neat! Executing the code given on this site, will result in a
-lower resolution of the field, because we overwrote `x_s` and `y_s` for the
-directional variogram estimation. In the example script, this is not the case
-and you will get a high resolution field.
-
-
-And Now for Some Cleanup
-^^^^^^^^^^^^^^^^^^^^^^^^
-
-In case you want all the downloaded data and scripts to be deleted, use
-following commands
-
-.. code-block:: python
-
- from shutil import rmtree
- os.remove('data.zip')
- os.remove('scripts.zip')
- rmtree('Herten-analog')
- rmtree('tools')
-
-And in case you want to play around a little bit more with the data, you can
-comment out the function calls ``download_herten()`` and
-``download_scripts()``, after they where called at least once and also comment
-out the cleanup. This way, the data will not be downloaded with every script
-execution.
-
-
-.. raw:: latex
-
- \clearpage
diff --git a/docs/source/tutorial_04_vec_field.rst b/docs/source/tutorial_04_vec_field.rst
deleted file mode 100644
index 8d251085..00000000
--- a/docs/source/tutorial_04_vec_field.rst
+++ /dev/null
@@ -1,98 +0,0 @@
-Tutorial 4: Random Vector Field Generation
-==========================================
-
-In 1970, Kraichnan was the first to suggest a randomization method.
-For studying the diffusion of single particles in a random incompressible
-velocity field, he came up with a randomization method which includes a
-projector which ensures the incompressibility of the vector field.
-
-
-Theoretical Background
-----------------------
-
-Without loss of generality we assume that the mean velocity :math:`\bar{U}` is oriented
-towards the direction of the first basis vector :math:`\mathbf{e}_1`. Our goal is now to
-generate random fluctuations with a given covariance model around this mean velocity.
-And at the same time, making sure that the velocity field remains incompressible or
-in other words, ensure :math:`\nabla \cdot \mathbf U = 0`.
-This can be done by using the randomization method we already know, but adding a
-projector to every mode being summed:
-
-
-.. math::
-
- \mathbf{U}(\mathbf{x}) = \bar{U} \mathbf{e}_1 - \sqrt{\frac{\sigma^{2}}{N}}
- \sum_{i=1}^{N} \mathbf{p}(\mathbf{k}_i) \left[ Z_{1,i}
- \cos\left( \langle \mathbf{k}_{i}, \mathbf{x} \rangle \right)
- + \sin\left( \langle \mathbf{k}_{i}, \mathbf{x} \rangle \right) \right]
-
-with the projector
-
-.. math::
-
- \mathbf{p}(\mathbf{k}_i) = \mathbf{e}_1 - \frac{\mathbf{k}_i k_1}{k^2} \; .
-
-By calculating :math:`\nabla \cdot \mathbf U = 0`, it can be verified, that
-the resulting field is indeed incompressible.
-
-
-Generating a Random Vector Field
---------------------------------
-
-As a first example we are going to generate a vector field with a Gaussian
-covariance model on a structured grid:
-
-.. code-block:: python
-
- import numpy as np
- import matplotlib.pyplot as plt
- from gstools import SRF, Gaussian
- x = np.arange(100)
- y = np.arange(100)
- model = Gaussian(dim=2, var=1, len_scale=10)
- srf = SRF(model, generator='VectorField')
- srf((x, y), mesh_type='structured', seed=19841203)
- srf.plot()
-
-And we get a beautiful streamflow plot:
-
-.. image:: pics/vec_srf_tut_gau.png
- :width: 600px
- :align: center
-
-Let us have a look at the influence of the covariance model. Choosing the
-exponential model and keeping all other parameters the same
-
-.. code-block:: python
-
- from gstools import Exponential
-
- model2 = Exponential(dim=2, var=1, len_scale=10)
- srf.model = model2
- srf((x, y), mesh_type='structured', seed=19841203)
- srf.plot()
-
-we get following result
-
-.. image:: pics/vec_srf_tut_exp.png
- :width: 600px
- :align: center
-
-and we see, that the wiggles are much "rougher" than the smooth Gaussian ones.
-
-
-Applications
-------------
-
-One great advantage of the Kraichnan method is, that after some initializations,
-one can compute the velocity field at arbitrary points, online, with hardly any
-overhead.
-This means, that for a Lagrangian transport simulation for example, the velocity
-can be evaluated at each particle position very efficiently and without any
-interpolation. These field interpolations are a common problem for Lagrangian
-methods.
-
-
-.. raw:: latex
-
- \clearpage
diff --git a/docs/source/tutorial_05_kriging.rst b/docs/source/tutorial_05_kriging.rst
deleted file mode 100755
index 7284b8b7..00000000
--- a/docs/source/tutorial_05_kriging.rst
+++ /dev/null
@@ -1,201 +0,0 @@
-Tutorial 5: Kriging
-===================
-
-The subpackage :py:mod:`gstools.krige` provides routines for Gaussian process regression, also known as kriging.
-Kriging is a method of data interpolation based on predefined covariance models.
-
-We provide two kinds of kriging routines:
-
-* Simple: The data is interpolated with a given mean value for the kriging field.
-* Ordinary: The mean of the resulting field is unkown and estimated during interpolation.
-
-
-Theoretical Background
-----------------------
-
-The aim of kriging is to derive the value of a field at some point :math:`x_0`,
-when there are fixed observed values :math:`z(x_1)\ldots z(x_n)` at given points :math:`x_i`.
-
-The resluting value :math:`z_0` at :math:`x_0` is calculated as a weighted mean:
-
-.. math::
-
- z_0 = \sum_{i=1}^n w_i \cdot z_i
-
-The weights :math:`W = (w_1,\ldots,w_n)` depent on the given covariance model and the location of the target point.
-
-The different kriging approaches provide different ways of calculating :math:`W`.
-
-
-Implementation
---------------
-
-The routines for kriging are almost identical to the routines for spatial random fields.
-First you define a covariance model, as described in :doc:`the SRF tutorial`,
-then you initialize the kriging class with this model:
-
-.. code-block:: python
-
- from gstools import Gaussian, krige
- # condtions
- cond_pos = ...
- cond_val = ...
- model = Gaussian(dim=1, var=0.5, len_scale=2)
- krig = krige.Simple(model, mean=1, cond_pos=cond_pos, cond_val=cond_val)
-
-The resulting field instance ``krig`` has the same methods as the :any:`SRF` class.
-You can call it to evaluate the kriged field at different points,
-you can plot the latest field or you can export the field and so on.
-Have a look at the documentation of :any:`Simple` and :any:`Ordinary`.
-
-
-Simple Kriging
---------------
-
-Simple kriging assumes a known mean of the data.
-For simplicity we assume a mean of 0,
-which can be achieved by subtracting the mean from the observed values and
-subsequently adding it to the resulting data.
-
-The resulting equation system for :math:`W` is given by:
-
-.. math::
-
- W = \begin{pmatrix}c(x_1,x_1) & \cdots & c(x_1,x_n) \\
- \vdots & \ddots & \vdots \\
- c(x_n,x_1) & \cdots & c(x_n,x_n)
- \end{pmatrix}^{-1}
- \begin{pmatrix}c(x_1,x_0) \\ \vdots \\ c(x_n,x_0) \end{pmatrix}
-
-Thereby :math:`c(x_i,x_j)` is the covariance of the given observations.
-
-
-Example
-^^^^^^^
-
-Here we use simple kriging in 1D (for plotting reasons) with 5 given observations/conditions.
-The mean of the field has to be given beforehand.
-
-.. code-block:: python
-
- import numpy as np
- from gstools import Gaussian, krige
- # condtions
- cond_pos = [0.3, 1.9, 1.1, 3.3, 4.7]
- cond_val = [0.47, 0.56, 0.74, 1.47, 1.74]
- # resulting grid
- gridx = np.linspace(0.0, 15.0, 151)
- # spatial random field class
- model = Gaussian(dim=1, var=0.5, len_scale=2)
- krig = krige.Simple(model, mean=1, cond_pos=cond_pos, cond_val=cond_val)
- krig(gridx)
- ax = krig.plot()
- ax.scatter(cond_pos, cond_val, color="k", zorder=10, label="Conditions")
- ax.legend()
-
-.. image:: pics/05_simple.png
- :width: 600px
- :align: center
-
-
-Ordinary Kriging
-----------------
-
-Ordinary kriging will estimate an appropriate mean of the field,
-based on the given observations/conditions and the covariance model used.
-
-The resulting system of equations for :math:`W` is given by:
-
-.. math::
-
- \begin{pmatrix}W\\\mu\end{pmatrix} = \begin{pmatrix}
- \gamma(x_1,x_1) & \cdots & \gamma(x_1,x_n) &1 \\
- \vdots & \ddots & \vdots & \vdots \\
- \gamma(x_n,x_1) & \cdots & \gamma(x_n,x_n) & 1 \\
- 1 &\cdots& 1 & 0
- \end{pmatrix}^{-1}
- \begin{pmatrix}\gamma(x_1,x_0) \\ \vdots \\ \gamma(x_n,x_0) \\ 1\end{pmatrix}
-
-Thereby :math:`\gamma(x_i,x_j)` is the semi-variogram of the given observations
-and :math:`\mu` is a Lagrange multiplier to minimize the kriging error and estimate the mean.
-
-
-Example
-^^^^^^^
-
-Here we use ordinary kriging in 1D (for plotting reasons) with 5 given observations/conditions.
-The estimated mean can be accessed by ``krig.mean``.
-
-.. code-block:: python
-
- import numpy as np
- from gstools import Gaussian, krige
- # condtions
- cond_pos = [0.3, 1.9, 1.1, 3.3, 4.7]
- cond_val = [0.47, 0.56, 0.74, 1.47, 1.74]
- # resulting grid
- gridx = np.linspace(0.0, 15.0, 151)
- # spatial random field class
- model = Gaussian(dim=1, var=0.5, len_scale=2)
- krig = krige.Ordinary(model, cond_pos=cond_pos, cond_val=cond_val)
- krig(gridx)
- ax = krig.plot()
- ax.scatter(cond_pos, cond_val, color="k", zorder=10, label="Conditions")
- ax.legend()
-
-.. image:: pics/05_ordinary.png
- :width: 600px
- :align: center
-
-
-Interface to PyKrige
---------------------
-
-To use fancier methods like `universal kriging `__,
-we provide an interface to `PyKrige `__.
-
-You can pass a GSTools Covariance Model to the PyKrige routines as ``variogram_model``.
-
-To demonstrate the general workflow, we compare the ordinary kriging of PyKrige
-with GSTools in 2D:
-
-.. code-block:: python
-
- import numpy as np
- from gstools import Gaussian, krige
- from pykrige.ok import OrdinaryKriging
- from matplotlib import pyplot as plt
-
- # conditioning data
- data = np.array([[0.3, 1.2, 0.47],
- [1.9, 0.6, 0.56],
- [1.1, 3.2, 0.74],
- [3.3, 4.4, 1.47],
- [4.7, 3.8, 1.74]])
- # grid definition for output field
- gridx = np.arange(0.0, 5.5, 0.1)
- gridy = np.arange(0.0, 6.5, 0.1)
- # a GSTools based covariance model
- cov_model = Gaussian(dim=2, len_scale=1, anis=.2, angles=-.5, var=.5, nugget=.1)
- # ordinary kriging with pykrige
- OK1 = OrdinaryKriging(data[:, 0], data[:, 1], data[:, 2], cov_model)
- z1, ss1 = OK1.execute('grid', gridx, gridy)
- plt.imshow(z1, origin="lower")
- plt.show()
- # ordinary kriging with gstools for comparison
- OK2 = krige.Ordinary(cov_model, [data[:, 0], data[:, 1]], data[:, 2])
- OK2.structured([gridx, gridy])
- OK2.plot()
-
-.. image:: pics/20_pykrige.png
- :width: 600px
- :align: center
-
-.. image:: pics/20_gstools.png
- :width: 600px
- :align: center
-
-
-.. raw:: latex
-
- \clearpage
diff --git a/docs/source/tutorial_06_conditioning.rst b/docs/source/tutorial_06_conditioning.rst
deleted file mode 100755
index 31ea93f0..00000000
--- a/docs/source/tutorial_06_conditioning.rst
+++ /dev/null
@@ -1,80 +0,0 @@
-Tutorial 6: Conditioned Fields
-==============================
-
-Kriged fields tend to approach the field mean outside the area of observations.
-To generate random fields, that coincide with given observations, but are still
-random according to a given covariance model away from the observations proximity,
-we provide the generation of conditioned random fields.
-
-
-Theoretical Background
-----------------------
-
-The idea behind conditioned random fields builds up on kriging.
-First we generate a field with a kriging method, then we generate a random field,
-and finally we generate another kriged field to eliminate the error between
-the random field and the kriged field of the given observations.
-
-To do so, you can choose between ordinary and simple kriging.
-In case of ordinary kriging, the mean of the SRF will be overwritten by the
-estimated mean.
-
-The setup of the spatial random field is the same as described in
-:doc:`the SRF tutorial`.
-You just need to add the conditions as described in :doc:`the kriging tutorial`:
-
-.. code-block:: python
-
- srf.set_condition(cond_pos, cond_val, "simple")
-
-or:
-
-.. code-block:: python
-
- srf.set_condition(cond_pos, cond_val, "ordinary")
-
-
-Example: Conditioning with Ordinary Kriging
--------------------------------------------
-
-Here we use ordinary kriging in 1D (for plotting reasons) with 5 given observations/conditions,
-to generate an ensemble of conditioned random fields.
-The estimated mean can be accessed by ``srf.mean``.
-
-.. code-block:: python
-
- import numpy as np
- from gstools import Gaussian, SRF
- import matplotlib.pyplot as plt
- # conditions
- cond_pos = [0.3, 1.9, 1.1, 3.3, 4.7]
- cond_val = [0.47, 0.56, 0.74, 1.47, 1.74]
- gridx = np.linspace(0.0, 15.0, 151)
- # spatial random field class
- model = Gaussian(dim=1, var=0.5, len_scale=2)
- srf = SRF(model)
- srf.set_condition(cond_pos, cond_val, "ordinary")
- fields = []
- for i in range(100):
- if i % 10 == 0: print(i)
- fields.append(srf(gridx, seed=i))
- label = "Conditioned ensemble" if i == 0 else None
- plt.plot(gridx, fields[i], color="k", alpha=0.1, label=label)
- plt.plot(gridx, np.full_like(gridx, srf.mean), label="estimated mean")
- plt.plot(gridx, np.mean(fields, axis=0), linestyle=':', label="Ensemble mean")
- plt.plot(gridx, srf.krige_field, linestyle='dashed', label="kriged field")
- plt.scatter(cond_pos, cond_val, color="k", zorder=10, label="Conditions")
- plt.legend()
- plt.show()
-
-.. image:: pics/06_ensemble.png
- :width: 600px
- :align: center
-
-As you can see, the kriging field coincides with the ensemble mean of the
-conditioned random fields and the estimated mean is the mean of the far-field.
-
-
-.. raw:: latex
-
- \clearpage
diff --git a/docs/source/tutorial_07_transformations.rst b/docs/source/tutorial_07_transformations.rst
deleted file mode 100755
index 6bea3b5c..00000000
--- a/docs/source/tutorial_07_transformations.rst
+++ /dev/null
@@ -1,188 +0,0 @@
-Tutorial 7: Field transformations
-=================================
-
-The generated fields of gstools are ordinary Gaussian random fields.
-In application there are several transformations to describe real world
-problems in an appropriate manner.
-
-GStools provides a submodule :py:mod:`gstools.transform` with a range of
-common transformations:
-
-.. currentmodule:: gstools.transform
-
-.. autosummary::
- binary
- boxcox
- zinnharvey
- normal_force_moments
- normal_to_lognormal
- normal_to_uniform
- normal_to_arcsin
- normal_to_uquad
-
-
-Implementation
---------------
-
-All the transformations take a field class, that holds a generated field,
-as input and will manipulate this field inplace.
-
-Simply import the transform submodule and apply a transformation to the srf class:
-
-.. code-block:: python
-
- from gstools import transform as tf
- ...
- tf.normal_to_lognormal(srf)
-
-
-In the following we will start from a simple random field following a Gaussian covariance:
-
-.. image:: pics/07_00_std.png
- :width: 600px
- :align: center
-
-
-1. Example: log-normal fields
------------------------------
-
-Here we transform a field to a log-normal distribution:
-
-.. code-block:: python
-
- from gstools import SRF, Gaussian
- from gstools import transform as tf
- # structured field with a size of 100x100 and a grid-size of 1x1
- x = y = range(100)
- model = Gaussian(dim=2, var=1, len_scale=10)
- srf = SRF(model, seed=20170519)
- srf.structured([x, y])
- tf.normal_to_lognormal(srf)
- srf.plot()
-
-
-.. image:: pics/07_01_lognormal.png
- :width: 600px
- :align: center
-
-
-2. Example: binary fields
--------------------------
-
-Here we transform a field to a binary field with only two values.
-The dividing value is the mean by default and the upper and lower values
-are derived to preserve the variance.
-
-.. code-block:: python
-
- from gstools import SRF, Gaussian
- from gstools import transform as tf
- # structured field with a size of 100x100 and a grid-size of 1x1
- x = y = range(100)
- model = Gaussian(dim=2, var=1, len_scale=10)
- srf = SRF(model, seed=20170519)
- srf.structured([x, y])
- tf.binary(srf)
- srf.plot()
-
-
-.. image:: pics/07_02_binary.png
- :width: 600px
- :align: center
-
-
-3. Example: Zinn & Harvey transformation
-----------------------------------------
-
-Here, we transform a field with the so called "Zinn & Harvey" transformation presented in
-`Zinn & Harvey (2003) `__.
-With this transformation, one could overcome the restriction that in ordinary
-Gaussian random fields the mean values are the ones being the most connected.
-
-.. code-block:: python
-
- from gstools import SRF, Gaussian
- from gstools import transform as tf
- # structured field with a size of 100x100 and a grid-size of 1x1
- x = y = range(100)
- model = Gaussian(dim=2, var=1, len_scale=10)
- srf = SRF(model, seed=20170519)
- srf.structured([x, y])
- tf.zinnharvey(srf, conn="high")
- srf.plot()
-
-
-.. image:: pics/07_03_zinnharvey.png
- :width: 600px
- :align: center
-
-
-4. Example: bimodal fields
---------------------------
-
-We provide two transformations to obtain bimodal distributions:
-
-* `arcsin `__.
-* `uquad `__.
-
-Both transformations will preserve the mean and variance of the given field by default.
-
-.. code-block:: python
-
- from gstools import SRF, Gaussian
- from gstools import transform as tf
- # structured field with a size of 100x100 and a grid-size of 1x1
- x = y = range(100)
- model = Gaussian(dim=2, var=1, len_scale=10)
- srf = SRF(model, seed=20170519)
- field = srf.structured([x, y])
- tf.normal_to_arcsin(srf)
- srf.plot()
-
-
-.. image:: pics/07_04_arcsin.png
- :width: 600px
- :align: center
-
-
-5. Example: Combinations
-------------------------
-
-You can combine different transformations simply by successively applying them.
-
-Here, we first force the single field realization to hold the given moments,
-namely mean and variance.
-Then we apply the Zinn & Harvey transformation to connect the low values.
-Afterwards the field is transformed to a binary field and last but not least,
-we transform it to log-values.
-
-
-.. code-block:: python
-
- from gstools import SRF, Gaussian
- from gstools import transform as tf
- # structured field with a size of 100x100 and a grid-size of 1x1
- x = y = range(100)
- model = Gaussian(dim=2, var=1, len_scale=10)
- srf = SRF(model, mean=-9, seed=20170519)
- srf.structured([x, y])
- tf.normal_force_moments(srf)
- tf.zinnharvey(srf, conn="low")
- tf.binary(srf)
- tf.normal_to_lognormal(srf)
- srf.plot()
-
-
-.. image:: pics/07_05_combine.png
- :width: 600px
- :align: center
-
-
-The resulting field could be interpreted as a transmissivity field, where
-the values of low permeability are the ones being the most connected
-and only two kinds of soil exist.
-
-
-.. raw:: latex
-
- \clearpage
diff --git a/docs/source/tutorials.rst b/docs/source/tutorials.rst
index e5b81501..5c2b6787 100644
--- a/docs/source/tutorials.rst
+++ b/docs/source/tutorials.rst
@@ -1,3 +1,5 @@
+.. _tutorials:
+
=================
GSTools Tutorials
=================
@@ -5,13 +7,16 @@ GSTools Tutorials
In the following you will find several Tutorials on how to use GSTools to
explore its whole beauty and power.
+
.. toctree::
+ :includehidden:
:maxdepth: 1
- tutorial_01_srf.rst
- tutorial_02_cov.rst
- tutorial_03_vario.rst
- tutorial_04_vec_field.rst
- tutorial_05_kriging.rst
- tutorial_06_conditioning.rst
- tutorial_07_transformations.rst
+ examples/01_random_field/index
+ examples/02_cov_model/index
+ examples/03_variogram/index
+ examples/04_vector_field/index
+ examples/05_kriging/index
+ examples/06_conditioned_fields/index
+ examples/07_transformations/index
+ examples/00_misc/index
diff --git a/examples/00_gaussian.py b/examples/00_gaussian.py
deleted file mode 100644
index 6800d18d..00000000
--- a/examples/00_gaussian.py
+++ /dev/null
@@ -1,8 +0,0 @@
-from gstools import SRF, Gaussian
-
-# structured field with a size of 100x100 and a grid-size of 1x1
-x = y = range(100)
-model = Gaussian(dim=2, var=1, len_scale=10)
-srf = SRF(model)
-srf.structured([x, y])
-srf.plot()
diff --git a/examples/00_misc/00_tpl_stable.py b/examples/00_misc/00_tpl_stable.py
new file mode 100644
index 00000000..0b8b8587
--- /dev/null
+++ b/examples/00_misc/00_tpl_stable.py
@@ -0,0 +1,60 @@
+r"""
+TPL Stable
+----------
+
+GSTools also implements truncated power law variograms,
+which can be represented as a superposition of scale dependant modes
+in form of standard variograms, which are truncated by
+a lower- :math:`\ell_{\mathrm{low}}` and
+an upper length-scale :math:`\ell_{\mathrm{up}}`.
+
+This example shows the truncated power law (:any:`TPLStable`) based on the
+:any:`Stable` covariance model and is given by
+
+.. math::
+ \gamma_{\ell_{\mathrm{low}},\ell_{\mathrm{up}}}(r) =
+ \intop_{\ell_{\mathrm{low}}}^{\ell_{\mathrm{up}}}
+ \gamma(r,\lambda) \frac{\rm d \lambda}{\lambda}
+
+with `Stable` modes on each scale:
+
+.. math::
+ \gamma(r,\lambda) &=
+ \sigma^2(\lambda)\cdot\left(1-
+ \exp\left[- \left(\frac{r}{\lambda}\right)^{\alpha}\right]
+ \right)\\
+ \sigma^2(\lambda) &= C\cdot\lambda^{2H}
+
+which gives Gaussian modes for ``alpha=2``
+or Exponential modes for ``alpha=1``.
+
+For :math:`\ell_{\mathrm{low}}=0` this results in:
+
+.. math::
+ \gamma_{\ell_{\mathrm{up}}}(r) &=
+ \sigma^2_{\ell_{\mathrm{up}}}\cdot\left(1-
+ \frac{2H}{\alpha} \cdot
+ E_{1+\frac{2H}{\alpha}}
+ \left[\left(\frac{r}{\ell_{\mathrm{up}}}\right)^{\alpha}\right]
+ \right) \\
+ \sigma^2_{\ell_{\mathrm{up}}} &=
+ C\cdot\frac{\ell_{\mathrm{up}}^{2H}}{2H}
+"""
+import numpy as np
+import gstools as gs
+
+x = y = np.linspace(0, 100, 100)
+model = gs.TPLStable(
+ dim=2, # spatial dimension
+ var=1, # variance (C is calculated internally, so variance is actually 1)
+ len_low=0, # lower truncation of the power law
+ len_scale=10, # length scale (a.k.a. range), len_up = len_low + len_scale
+ nugget=0.1, # nugget
+ anis=0.5, # anisotropy between main direction and transversal ones
+ angles=np.pi / 4, # rotation angles
+ alpha=1.5, # shape parameter from the stable model
+ hurst=0.7, # hurst coefficient from the power law
+)
+srf = gs.SRF(model, mean=1.0, seed=19970221)
+srf.structured([x, y])
+srf.plot()
diff --git a/examples/00_misc/01_export.py b/examples/00_misc/01_export.py
new file mode 100644
index 00000000..cbc89706
--- /dev/null
+++ b/examples/00_misc/01_export.py
@@ -0,0 +1,12 @@
+"""
+Export
+------
+"""
+
+import gstools as gs
+
+x = y = range(100)
+model = gs.Gaussian(dim=2, var=1, len_scale=10)
+srf = gs.SRF(model)
+field = srf((x, y), mesh_type="structured")
+srf.vtk_export(filename="field")
diff --git a/examples/19_check_rand_meth_sampling.py b/examples/00_misc/02_check_rand_meth_sampling.py
similarity index 90%
rename from examples/19_check_rand_meth_sampling.py
rename to examples/00_misc/02_check_rand_meth_sampling.py
index 2ea5d5ed..c03213c6 100644
--- a/examples/19_check_rand_meth_sampling.py
+++ b/examples/00_misc/02_check_rand_meth_sampling.py
@@ -1,8 +1,11 @@
+"""
+Check Random Sampling
+---------------------
+"""
import numpy as np
from matplotlib import pyplot as plt
from mpl_toolkits.mplot3d import Axes3D
-
-from gstools import SRF, Stable
+import gstools as gs
def norm_rad(vec):
@@ -30,7 +33,6 @@ def plot_rand_meth_samples(generator):
z = np.outer(np.ones(np.size(u)), np.cos(v))
ax.plot_surface(x, y, z, rstride=4, cstride=4, color="b", alpha=0.1)
ax.scatter(norm[0], norm[1], norm[2])
- ax.set_aspect("equal")
elif generator.model.dim == 2:
ax = fig.add_subplot(121)
u = np.linspace(0, 2 * np.pi, 100)
@@ -48,17 +50,17 @@ def plot_rand_meth_samples(generator):
ax.set_title("Direction sampling")
ax = fig.add_subplot(122)
- # x = np.linspace(0, np.max(rad))
x = np.linspace(0, 10 / generator.model.integral_scale)
y = generator.model.spectral_rad_pdf(x)
- ax.plot(x, y)
+ ax.plot(x, y, label="radial spectral density")
sample_in = np.sum(rad <= np.max(x))
ax.hist(rad[rad <= np.max(x)], bins=sample_in // 50, density=True)
ax.set_xlim([0, np.max(x)])
ax.set_title("Radius samples shown {}/{}".format(sample_in, len(rad)))
+ ax.legend()
fig.show()
-model = Stable(dim=3, alpha=1.5)
-srf = SRF(model)
+model = gs.Stable(dim=3, alpha=1.5)
+srf = gs.SRF(model, seed=2020)
plot_rand_meth_samples(srf.generator)
diff --git a/examples/00_misc/README.rst b/examples/00_misc/README.rst
new file mode 100644
index 00000000..cf021250
--- /dev/null
+++ b/examples/00_misc/README.rst
@@ -0,0 +1,7 @@
+Miscellaneous
+=============
+
+A few miscellaneous examples
+
+Gallery
+-------
diff --git a/examples/01_random_field/00_gaussian.py b/examples/01_random_field/00_gaussian.py
new file mode 100644
index 00000000..726ad653
--- /dev/null
+++ b/examples/01_random_field/00_gaussian.py
@@ -0,0 +1,38 @@
+"""
+A Very Simple Example
+---------------------
+
+We are going to start with a very simple example of a spatial random field
+with an isotropic Gaussian covariance model and following parameters:
+
+- variance :math:`\sigma^2=1`
+- correlation length :math:`\lambda=10`
+
+First, we set things up and create the axes for the field. We are going to
+need the :any:`SRF` class for the actual generation of the spatial random field.
+But :any:`SRF` also needs a covariance model and we will simply take the
+:any:`Gaussian` model.
+"""
+
+import gstools as gs
+
+x = y = range(100)
+
+###############################################################################
+# Now we create the covariance model with the parameters :math:`\sigma^2` and
+# :math:`\lambda` and hand it over to :any:`SRF`. By specifying a seed,
+# we make sure to create reproducible results:
+
+model = gs.Gaussian(dim=2, var=1, len_scale=10)
+srf = gs.SRF(model, seed=20170519)
+
+###############################################################################
+# With these simple steps, everything is ready to create our first random field.
+# We will create the field on a structured grid (as you might have guessed from
+# the `x` and `y`), which makes it easier to plot.
+
+field = srf.structured([x, y])
+srf.plot()
+
+###############################################################################
+# Wow, that was pretty easy!
diff --git a/examples/01_random_field/01_srf_ensemble.py b/examples/01_random_field/01_srf_ensemble.py
new file mode 100644
index 00000000..6174d4ce
--- /dev/null
+++ b/examples/01_random_field/01_srf_ensemble.py
@@ -0,0 +1,52 @@
+"""
+Creating an Ensemble of Fields
+------------------------------
+
+Creating an ensemble of random fields would also be
+a great idea. Let's reuse most of the previous code.
+"""
+
+import numpy as np
+import matplotlib.pyplot as pt
+import gstools as gs
+
+x = y = np.arange(100)
+
+model = gs.Gaussian(dim=2, var=1, len_scale=10)
+srf = gs.SRF(model)
+
+###############################################################################
+# This time, we did not provide a seed to :any:`SRF`, as the seeds will used
+# during the actual computation of the fields. We will create four ensemble
+# members, for better visualisation and save them in a list and in a first
+# step, we will be using the loop counter as the seeds.
+
+
+ens_no = 4
+field = []
+for i in range(ens_no):
+ field.append(srf.structured([x, y], seed=i))
+
+###############################################################################
+# Now let's have a look at the results:
+
+fig, ax = pt.subplots(2, 2, sharex=True, sharey=True)
+ax = ax.flatten()
+for i in range(ens_no):
+ ax[i].imshow(field[i].T, origin="lower")
+pt.show()
+
+###############################################################################
+# Using better Seeds
+# ^^^^^^^^^^^^^^^^^^
+#
+# It is not always a good idea to use incrementing seeds. Therefore GSTools
+# provides a seed generator :any:`MasterRNG`. The loop, in which the fields are
+# generated would then look like
+
+
+from gstools.random import MasterRNG
+
+seed = MasterRNG(20170519)
+for i in range(ens_no):
+ field.append(srf.structured([x, y], seed=seed()))
diff --git a/examples/01_random_field/02_fancier.py b/examples/01_random_field/02_fancier.py
new file mode 100644
index 00000000..f27dccde
--- /dev/null
+++ b/examples/01_random_field/02_fancier.py
@@ -0,0 +1,30 @@
+"""
+Creating Fancier Fields
+-----------------------
+
+Only using Gaussian covariance fields gets boring. Now we are going to create
+much rougher random fields by using an exponential covariance model and we are going to make them anisotropic.
+
+The code is very similar to the previous examples, but with a different
+covariance model class :any:`Exponential`. As model parameters we a using
+following
+
+- variance :math:`\sigma^2=1`
+- correlation length :math:`\lambda=(12, 3)^T`
+- rotation angle :math:`\theta=\pi/8`
+
+"""
+
+import numpy as np
+import gstools as gs
+
+x = y = np.arange(100)
+model = gs.Exponential(dim=2, var=1, len_scale=[12.0, 3.0], angles=np.pi / 8)
+srf = gs.SRF(model, seed=20170519)
+srf.structured([x, y])
+srf.plot()
+
+###############################################################################
+# The anisotropy ratio could also have been set with
+
+model = gs.Exponential(dim=2, var=1, len_scale=12, anis=0.25, angles=np.pi / 8)
diff --git a/examples/01_random_field/03_unstr_srf_export.py b/examples/01_random_field/03_unstr_srf_export.py
new file mode 100644
index 00000000..a43c00eb
--- /dev/null
+++ b/examples/01_random_field/03_unstr_srf_export.py
@@ -0,0 +1,32 @@
+"""
+Using an Unstructured Grid
+--------------------------
+
+For many applications, the random fields are needed on an unstructured grid.
+Normally, such a grid would be read in, but we can simply generate one and
+then create a random field at those coordinates.
+"""
+import numpy as np
+import gstools as gs
+
+###############################################################################
+# Creating our own unstructured grid
+seed = gs.random.MasterRNG(19970221)
+rng = np.random.RandomState(seed())
+x = rng.randint(0, 100, size=10000)
+y = rng.randint(0, 100, size=10000)
+
+model = gs.Exponential(dim=2, var=1, len_scale=[12, 3], angles=np.pi / 8)
+srf = gs.SRF(model, seed=20170519)
+field = srf((x, y))
+srf.vtk_export("field")
+# Or create a PyVista dataset
+# mesh = srf.to_pyvista()
+
+###############################################################################
+ax = srf.plot()
+ax.set_aspect("equal")
+
+###############################################################################
+# Comparing this image to the previous one, you can see that be using the same
+# seed, the same field can be computed on different grids.
diff --git a/examples/01_random_field/04_srf_merge.py b/examples/01_random_field/04_srf_merge.py
new file mode 100644
index 00000000..366558da
--- /dev/null
+++ b/examples/01_random_field/04_srf_merge.py
@@ -0,0 +1,46 @@
+"""
+Merging two Fields
+------------------
+
+We can even generate the same field realisation on different grids. Let's try
+to merge two unstructured rectangular fields.
+
+"""
+import numpy as np
+import gstools as gs
+
+# creating our own unstructured grid
+seed = gs.random.MasterRNG(19970221)
+rng = np.random.RandomState(seed())
+x = rng.randint(0, 100, size=10000)
+y = rng.randint(0, 100, size=10000)
+
+model = gs.Exponential(dim=2, var=1, len_scale=[12, 3], angles=np.pi / 8)
+srf = gs.SRF(model, seed=20170519)
+field1 = srf((x, y))
+srf.plot()
+###############################################################################
+# But now we extend the field on the right hand side by creating a new
+# unstructured grid and calculating a field with the same parameters and the
+# same seed on it:
+
+# new grid
+seed = gs.random.MasterRNG(20011012)
+rng = np.random.RandomState(seed())
+x2 = rng.randint(99, 150, size=10000)
+y2 = rng.randint(20, 80, size=10000)
+
+field2 = srf((x2, y2))
+ax = srf.plot()
+ax.tricontourf(x, y, field1.T, levels=256)
+ax.set_aspect("equal")
+
+###############################################################################
+# The slight mismatch where the two fields were merged is merely due to
+# interpolation problems of the plotting routine. You can convince yourself
+# be increasing the resolution of the grids by a factor of 10.
+#
+# Of course, this merging could also have been done by appending the grid
+# point ``(x2, y2)`` to the original grid ``(x, y)`` before generating the field.
+# But one application scenario would be to generate hugh fields, which would not
+# fit into memory anymore.
diff --git a/examples/01_random_field/README.rst b/examples/01_random_field/README.rst
new file mode 100644
index 00000000..a0452568
--- /dev/null
+++ b/examples/01_random_field/README.rst
@@ -0,0 +1,17 @@
+Tutorial 1: Random Field Generation
+===================================
+
+The main feature of GSTools is the spatial random field generator :any:`SRF`,
+which can generate random fields following a given covariance model.
+The generator provides a lot of nice features, which will be explained in
+the following
+
+GSTools generates spatial random fields with a given covariance model or
+semi-variogram. This is done by using the so-called randomization method.
+The spatial random field is represented by a stochastic Fourier integral
+and its discretised modes are evaluated at random frequencies.
+
+GSTools supports arbitrary and non-isotropic covariance models.
+
+Gallery
+-------
diff --git a/examples/01_tpl_stable.py b/examples/01_tpl_stable.py
deleted file mode 100644
index 1e7fae1d..00000000
--- a/examples/01_tpl_stable.py
+++ /dev/null
@@ -1,18 +0,0 @@
-import numpy as np
-from gstools import SRF, TPLStable
-
-x = y = np.linspace(0, 100, 100)
-model = TPLStable(
- dim=2, # spatial dimension
- var=1, # variance (C is calculated internally, so that the variance is actually 1)
- len_low=0, # lower truncation of the power law
- len_scale=10, # length scale (a.k.a. range), len_up = len_low + len_scale
- nugget=0.1, # nugget
- anis=0.5, # anisotropy between main direction and transversal ones
- angles=np.pi / 4, # rotation angles
- alpha=1.5, # shape parameter from the stable model
- hurst=0.7, # hurst coefficient from the power law
-)
-srf = SRF(model, mean=1, mode_no=1000, seed=19970221, verbose=True)
-srf.structured([x, y])
-srf.plot()
diff --git a/examples/02_cov_model/00_intro.py b/examples/02_cov_model/00_intro.py
new file mode 100644
index 00000000..55d18fdc
--- /dev/null
+++ b/examples/02_cov_model/00_intro.py
@@ -0,0 +1,74 @@
+"""
+Introductory example
+====================
+
+Let us start with a short example of a self defined model (Of course, we
+provide a lot of predefined models [See: :any:`gstools.covmodel`],
+but they all work the same way).
+Therefore we reimplement the Gaussian covariance model
+by defining just the "normalized"
+`correlation `_
+function:
+"""
+
+import numpy as np
+import gstools as gs
+
+
+# use CovModel as the base-class
+class Gau(gs.CovModel):
+ def cor(self, h):
+ return np.exp(-h ** 2)
+
+
+###############################################################################
+# Here the parameter ``h`` stands for the normalized range ``r / len_scale``.
+# Now we can instantiate this model:
+
+model = Gau(dim=2, var=2.0, len_scale=10)
+
+###############################################################################
+# To have a look at the variogram, let's plot it:
+
+model.plot()
+
+###############################################################################
+# This is almost identical to the already provided :any:`Gaussian` model.
+# There, a scaling factor is implemented so the len_scale coincides with the
+# integral scale:
+
+gau_model = gs.Gaussian(dim=2, var=2.0, len_scale=10)
+gau_model.plot()
+
+
+###############################################################################
+# Parameters
+# ----------
+#
+# We already used some parameters, which every covariance models has.
+# The basic ones are:
+#
+# - **dim** : dimension of the model
+# - **var** : variance of the model (on top of the subscale variance)
+# - **len_scale** : length scale of the model
+# - **nugget** : nugget (subscale variance) of the model
+#
+# These are the common parameters used to characterize
+# a covariance model and are therefore used by every model in GSTools.
+# You can also access and reset them:
+
+print("old model:", model)
+model.dim = 3
+model.var = 1
+model.len_scale = 15
+model.nugget = 0.1
+print("new model:", model)
+
+
+###############################################################################
+# .. note::
+#
+# - The sill of the variogram is calculated by ``sill = variance + nugget``
+# So we treat the variance as everything **above** the nugget,
+# which is sometimes called **partial sill**.
+# - A covariance model can also have additional parameters.
diff --git a/examples/02_cov_model/01_basic_methods.py b/examples/02_cov_model/01_basic_methods.py
new file mode 100755
index 00000000..fb9bfe7a
--- /dev/null
+++ b/examples/02_cov_model/01_basic_methods.py
@@ -0,0 +1,44 @@
+r"""
+Basic Methods
+=============
+
+The covariance model class :any:`CovModel` of GSTools provides a set of handy
+methods.
+
+One of the following functions defines the main characterization of the
+variogram:
+
+- ``CovModel.variogram`` : The variogram of the model given by
+
+ .. math::
+ \gamma\left(r\right)=
+ \sigma^2\cdot\left(1-\rho\left(r\right)\right)+n
+
+- ``CovModel.covariance`` : The (auto-)covariance of the model given by
+
+ .. math::
+ C\left(r\right)= \sigma^2\cdot\rho\left(r\right)
+
+- ``CovModel.correlation`` : The (auto-)correlation
+ (or normalized covariance) of the model given by
+
+ .. math::
+ \rho\left(r\right)
+
+- ``CovModel.cor`` : The normalized correlation taking a
+ normalized range given by:
+
+ .. math::
+ \mathrm{cor}\left(\frac{r}{\ell}\right) = \rho\left(r\right)
+
+
+As you can see, it is the easiest way to define a covariance model by giving a
+correlation function as demonstrated in the introductory example.
+If one of the above functions is given, the others will be determined:
+"""
+import gstools as gs
+
+model = gs.Exponential(dim=3, var=2.0, len_scale=10, nugget=0.5)
+ax = model.plot("variogram")
+model.plot("covariance", ax=ax)
+model.plot("correlation", ax=ax)
diff --git a/examples/02_cov_model/02_aniso_rotation.py b/examples/02_cov_model/02_aniso_rotation.py
new file mode 100755
index 00000000..58d7b736
--- /dev/null
+++ b/examples/02_cov_model/02_aniso_rotation.py
@@ -0,0 +1,54 @@
+"""
+Anisotropy and Rotation
+=======================
+
+The internally used (semi-) variogram
+represents the isotropic case for the model.
+Nevertheless, you can provide anisotropy ratios by:
+"""
+import gstools as gs
+
+model = gs.Gaussian(dim=3, var=2.0, len_scale=10, anis=0.5)
+print(model.anis)
+print(model.len_scale_vec)
+
+
+###############################################################################
+# As you can see, we defined just one anisotropy-ratio
+# and the second transversal direction was filled up with ``1.``.
+# You can get the length-scales in each direction by
+# the attribute :any:`CovModel.len_scale_vec`. For full control you can set
+# a list of anistropy ratios: ``anis=[0.5, 0.4]``.
+#
+# Alternatively you can provide a list of length-scales:
+
+model = gs.Gaussian(dim=3, var=2.0, len_scale=[10, 5, 4])
+model.plot("vario_spatial")
+print("Anisotropy representations:")
+print("Anis. ratios:", model.anis)
+print("Main length scale", model.len_scale)
+print("All length scales", model.len_scale_vec)
+
+
+###############################################################################
+# Rotation Angles
+# ---------------
+#
+# The main directions of the field don't have to coincide with the spatial
+# directions :math:`x`, :math:`y` and :math:`z`. Therefore you can provide
+# rotation angles for the model:
+
+model = gs.Gaussian(dim=3, var=2.0, len_scale=[10, 2], angles=2.5)
+model.plot("vario_spatial")
+print("Rotation angles", model.angles)
+
+###############################################################################
+# Again, the angles were filled up with ``0.`` to match the dimension and you
+# could also provide a list of angles. The number of angles depends on the
+# given dimension:
+#
+# - in 1D: no rotation performable
+# - in 2D: given as rotation around z-axis
+# - in 3D: given by yaw, pitch, and roll (known as
+# `Tait–Bryan `_
+# angles)
diff --git a/examples/02_cov_model/03_spectral_methods.py b/examples/02_cov_model/03_spectral_methods.py
new file mode 100755
index 00000000..677811a9
--- /dev/null
+++ b/examples/02_cov_model/03_spectral_methods.py
@@ -0,0 +1,46 @@
+r"""
+Spectral methods
+================
+
+The spectrum of a covariance model is given by:
+
+.. math:: S(\mathbf{k}) = \left(\frac{1}{2\pi}\right)^n
+ \int C(\Vert\mathbf{r}\Vert) e^{i b\mathbf{k}\cdot\mathbf{r}} d^n\mathbf{r}
+
+Since the covariance function :math:`C(r)` is radially symmetric, we can
+calculate this by the
+`hankel-transformation `_:
+
+.. math:: S(k) = \left(\frac{1}{2\pi}\right)^n \cdot
+ \frac{(2\pi)^{n/2}}{(bk)^{n/2-1}}
+ \int_0^\infty r^{n/2-1} C(r) J_{n/2-1}(bkr) r dr
+
+Where :math:`k=\left\Vert\mathbf{k}\right\Vert`.
+
+Depending on the spectrum, the spectral-density is defined by:
+
+.. math:: \tilde{S}(k) = \frac{S(k)}{\sigma^2}
+
+You can access these methods by:
+"""
+import gstools as gs
+
+model = gs.Gaussian(dim=3, var=2.0, len_scale=10)
+ax = model.plot("spectrum")
+model.plot("spectral_density", ax=ax)
+
+###############################################################################
+# .. note::
+# The spectral-density is given by the radius of the input phase. But it is
+# **not** a probability density function for the radius of the phase.
+# To obtain the pdf for the phase-radius, you can use the methods
+# :any:`CovModel.spectral_rad_pdf`
+# or :any:`CovModel.ln_spectral_rad_pdf` for the logarithm.
+#
+# The user can also provide a cdf (cumulative distribution function) by
+# defining a method called ``spectral_rad_cdf``
+# and/or a ppf (percent-point function)
+# by ``spectral_rad_ppf``.
+#
+# The attributes :any:`CovModel.has_cdf`
+# and :any:`CovModel.has_ppf` will check for that.
diff --git a/examples/02_cov_model/04_different_scales.py b/examples/02_cov_model/04_different_scales.py
new file mode 100755
index 00000000..cd6f4dee
--- /dev/null
+++ b/examples/02_cov_model/04_different_scales.py
@@ -0,0 +1,68 @@
+r"""
+Different scales
+================
+
+Besides the length-scale, there are many other ways of characterizing a certain
+scale of a covariance model. We provide two common scales with the covariance
+model.
+
+Integral scale
+--------------
+
+The `integral scale `_
+of a covariance model is calculated by:
+
+.. math:: I = \int_0^\infty \rho(r) dr
+
+You can access it by:
+"""
+import gstools as gs
+
+model = gs.Stable(dim=3, var=2.0, len_scale=10)
+print("Main integral scale:", model.integral_scale)
+print("All integral scales:", model.integral_scale_vec)
+
+
+###############################################################################
+# You can also specify integral length scales like the ordinary length scale,
+# and len_scale/anis will be recalculated:
+
+model = gs.Stable(dim=3, var=2.0, integral_scale=[10, 4, 2])
+print("Anisotropy ratios:", model.anis)
+print("Main length scale:", model.len_scale)
+print("All length scales:", model.len_scale_vec)
+print("Main integral scale:", model.integral_scale)
+print("All integral scales:", model.integral_scale_vec)
+
+
+###############################################################################
+# Percentile scale
+# ----------------
+#
+# Another scale characterizing the covariance model, is the percentile scale.
+# It is the distance, where the normalized
+# variogram reaches a certain percentage of its sill.
+
+model = gs.Stable(dim=3, var=2.0, len_scale=10)
+per_scale = model.percentile_scale(0.9)
+int_scale = model.integral_scale
+len_scale = model.len_scale
+print("90% Percentile scale:", per_scale)
+print("Integral scale:", int_scale)
+print("Length scale:", len_scale)
+
+###############################################################################
+# .. note::
+#
+# The nugget is neglected by the percentile scale.
+#
+#
+# Comparison
+# ----------
+
+ax = model.plot()
+ax.axhline(1.8, color="k", label=r"90% percentile")
+ax.axvline(per_scale, color="k", linestyle="--", label=r"90% percentile scale")
+ax.axvline(int_scale, color="k", linestyle="-.", label=r"integral scale")
+ax.axvline(len_scale, color="k", linestyle=":", label=r"length scale")
+ax.legend()
diff --git a/examples/02_cov_model/05_additional_para.py b/examples/02_cov_model/05_additional_para.py
new file mode 100755
index 00000000..dc9012a2
--- /dev/null
+++ b/examples/02_cov_model/05_additional_para.py
@@ -0,0 +1,45 @@
+r"""
+Additional Parameters
+=====================
+
+Let's pimp our self-defined model ``Gau`` from the introductory example
+by setting the exponent as an additional parameter:
+
+.. math::
+ \rho(r) := \exp\left(-\left(\frac{r}{\ell}\right)^{\alpha}\right)
+
+This leads to the so called **stable** covariance model and we can define it by
+"""
+import numpy as np
+import gstools as gs
+
+
+class Stab(gs.CovModel):
+ def default_opt_arg(self):
+ return {"alpha": 1.5}
+
+ def cor(self, h):
+ return np.exp(-h ** self.alpha)
+
+
+###############################################################################
+# As you can see, we override the method :any:`CovModel.default_opt_arg`
+# to provide a standard value for the optional argument ``alpha``.
+# We can access it in the correlation function by ``self.alpha``
+#
+# Now we can instantiate this model by either setting alpha implicitly with
+# the default value or explicitly:
+
+model1 = Stab(dim=2, var=2.0, len_scale=10)
+model2 = Stab(dim=2, var=2.0, len_scale=10, alpha=0.5)
+ax = model1.plot()
+model2.plot(ax=ax)
+
+###############################################################################
+# Apparently, the parameter alpha controls the slope of the variogram
+# and consequently the roughness of a generated random field.
+#
+# .. note::
+#
+# You don't have to override the :any:`CovModel.default_opt_arg`,
+# but you will get a ValueError if you don't set it on creation.
diff --git a/examples/02_cov_model/06_fitting_para_ranges.py b/examples/02_cov_model/06_fitting_para_ranges.py
new file mode 100755
index 00000000..f73253ce
--- /dev/null
+++ b/examples/02_cov_model/06_fitting_para_ranges.py
@@ -0,0 +1,74 @@
+"""
+Fitting variogram data
+======================
+
+The model class comes with a routine to fit the model-parameters to given
+variogram data. In the following we will use the self defined stable model
+from a previous example.
+"""
+import numpy as np
+import gstools as gs
+
+
+class Stab(gs.CovModel):
+ def default_opt_arg(self):
+ return {"alpha": 1.5}
+
+ def cor(self, h):
+ return np.exp(-h ** self.alpha)
+
+
+# Exemplary variogram data (e.g. estimated from field observations)
+bins = [1.0, 3.0, 5.0, 7.0, 9.0, 11.0]
+est_vario = [0.2, 0.5, 0.6, 0.8, 0.8, 0.9]
+# fitting model
+model = Stab(dim=2)
+# we have to provide boundaries for the parameters
+model.set_arg_bounds(alpha=[0, 3])
+results, pcov = model.fit_variogram(bins, est_vario, nugget=False)
+print("Results:", results)
+
+###############################################################################
+
+ax = model.plot()
+ax.scatter(bins, est_vario, color="k", label="sample variogram")
+ax.legend()
+
+
+###############################################################################
+# As you can see, we have to provide boundaries for the parameters.
+# As a default, the following bounds are set:
+#
+# - additional parameters: ``[-np.inf, np.inf]``
+# - variance: ``[0.0, np.inf]``
+# - len_scale: ``[0.0, np.inf]``
+# - nugget: ``[0.0, np.inf]``
+#
+# Also, you can deselect parameters from fitting, so their predefined values
+# will be kept. In our case, we fixed a ``nugget`` of ``0.0``, which was set
+# by default. You can deselect any standard or
+# optional argument of the covariance model.
+# The second return value ``pcov`` is the estimated covariance of ``popt`` from
+# the used scipy routine :any:`scipy.optimize.curve_fit`.
+#
+# You can use the following methods to manipulate the used bounds:
+#
+# .. currentmodule:: gstools.covmodel
+#
+# .. autosummary::
+# CovModel.default_opt_arg_bounds
+# CovModel.default_arg_bounds
+# CovModel.set_arg_bounds
+# CovModel.check_arg_bounds
+#
+# You can override the :any:`CovModel.default_opt_arg_bounds`
+# to provide standard bounds for your additional parameters.
+#
+# To access the bounds you can use:
+#
+# .. autosummary::
+# CovModel.var_bounds
+# CovModel.len_scale_bounds
+# CovModel.nugget_bounds
+# CovModel.opt_arg_bounds
+# CovModel.arg_bounds
diff --git a/examples/02_cov_model/README.rst b/examples/02_cov_model/README.rst
new file mode 100644
index 00000000..6ea0032a
--- /dev/null
+++ b/examples/02_cov_model/README.rst
@@ -0,0 +1,67 @@
+.. _tutorial_02_cov:
+
+Tutorial 2: The Covariance Model
+================================
+
+One of the core-features of GSTools is the powerful :any:`CovModel`
+class, which allows you to easily define arbitrary covariance models by
+yourself. The resulting models provide a bunch of nice features to explore the
+covariance models.
+
+
+A covariance model is used to characterize the
+`semi-variogram `_,
+denoted by :math:`\gamma`, of a spatial random field.
+In GSTools, we use the following form for an isotropic and stationary field:
+
+.. math::
+ \gamma\left(r\right)=
+ \sigma^2\cdot\left(1-\rho\left(r\right)\right)+n
+
+Where:
+
+ - :math:`\rho(r)` is the so called
+ `correlation `_
+ function depending on the distance :math:`r`
+ - :math:`\sigma^2` is the variance
+ - :math:`n` is the nugget (subscale variance)
+
+.. note::
+
+ We are not limited to isotropic models. GSTools supports anisotropy ratios
+ for length scales in orthogonal transversal directions like:
+
+ - :math:`x` (main direction)
+ - :math:`y` (1. transversal direction)
+ - :math:`z` (2. transversal direction)
+
+ These main directions can also be rotated.
+ Just have a look at the corresponding examples.
+
+Provided Covariance Models
+--------------------------
+
+.. currentmodule:: gstools.covmodel
+
+The following standard covariance models are provided by GSTools
+
+.. autosummary::
+ Gaussian
+ Exponential
+ Matern
+ Stable
+ Rational
+ Linear
+ Circular
+ Spherical
+ Intersection
+
+As a special feature, we also provide truncated power law (TPL) covariance models
+
+.. autosummary::
+ TPLGaussian
+ TPLExponential
+ TPLStable
+
+Gallery
+-------
diff --git a/examples/02_fit_variogram.py b/examples/02_fit_variogram.py
deleted file mode 100644
index 3a36a9ba..00000000
--- a/examples/02_fit_variogram.py
+++ /dev/null
@@ -1,19 +0,0 @@
-import numpy as np
-from gstools import SRF, Exponential, Stable, vario_estimate_unstructured
-
-# generate a synthetic field with an exponential model
-x = np.random.RandomState(19970221).rand(1000) * 100.0
-y = np.random.RandomState(20011012).rand(1000) * 100.0
-model = Exponential(dim=2, var=2, len_scale=8)
-srf = SRF(model, mean=0, seed=19970221)
-field = srf((x, y))
-# estimate the variogram of the field with 40 bins
-bins = np.arange(40)
-bin_center, gamma = vario_estimate_unstructured((x, y), field, bins)
-# fit the variogram with a stable model. (no nugget fitted)
-fit_model = Stable(dim=2)
-fit_model.fit_variogram(bin_center, gamma, nugget=False)
-# output
-ax = fit_model.plot(x_max=40)
-ax.plot(bin_center, gamma)
-print(fit_model)
diff --git a/examples/03_cov_model.py b/examples/03_cov_model.py
deleted file mode 100644
index 9101c360..00000000
--- a/examples/03_cov_model.py
+++ /dev/null
@@ -1,81 +0,0 @@
-from gstools import CovModel
-import numpy as np
-
-# use CovModel as the base-class
-class Gau(CovModel):
- def correlation(self, r):
- return np.exp(-(r / self.len_scale) ** 2)
-
-
-model = Gau(dim=2, var=2.0, len_scale=10)
-
-model.plot()
-
-print(model.dim, model.var, model.len_scale, model.nugget, model.sill)
-model.dim = 3
-model.var = 1
-model.len_scale = 15
-model.nugget = 0.1
-print(model.dim, model.var, model.len_scale, model.nugget, model.sill)
-
-model = Gau(dim=3, var=2.0, len_scale=10, anis=0.5)
-print(model.anis)
-print(model.len_scale_vec)
-
-model = Gau(dim=3, var=2.0, len_scale=[10, 5, 4])
-print(model.anis)
-print(model.len_scale)
-print(model.len_scale_vec)
-
-model = Gau(dim=3, var=2.0, len_scale=10, angles=2.5)
-print(model.angles)
-
-model = Gau(dim=3, var=2.0, len_scale=10, nugget=0.5)
-print(model.variogram(10.0))
-print(model.covariance(10.0))
-print(model.correlation(10.0))
-
-model = Gau(dim=3, var=2.0, len_scale=10)
-print(model.spectrum(0.1))
-print(model.spectral_density(0.1))
-
-model = Gau(dim=3, var=2.0, len_scale=10)
-print(model.integral_scale)
-print(model.integral_scale_vec)
-
-model = Gau(dim=3, var=2.0, integral_scale=[10, 4, 2])
-print(model.anis)
-print(model.len_scale)
-print(model.len_scale_vec)
-print(model.integral_scale)
-print(model.integral_scale_vec)
-
-model = Gau(dim=3, var=2.0, len_scale=10)
-print(model.percentile_scale(0.9))
-
-
-class Stab(CovModel):
- def default_opt_arg(self):
- return {"alpha": 1.5}
-
- def correlation(self, r):
- return np.exp(-(r / self.len_scale) ** self.alpha)
-
-
-model1 = Stab(dim=2, var=2.0, len_scale=10)
-model2 = Stab(dim=2, var=2.0, len_scale=10, alpha=0.5)
-print(model1)
-print(model2)
-
-# data
-x = [1.0, 3.0, 5.0, 7.0, 9.0, 11.0]
-y = [0.2, 0.5, 0.6, 0.8, 0.8, 0.9]
-# fitting model
-model = Stab(dim=2)
-# we have to provide boundaries for the parameters
-model.set_arg_bounds(alpha=[0, 3])
-results, pcov = model.fit_variogram(x, y, nugget=False)
-print(results)
-
-ax = model.plot()
-ax.scatter(x, y, color="k")
diff --git a/examples/03_variogram/00_fit_variogram.py b/examples/03_variogram/00_fit_variogram.py
new file mode 100644
index 00000000..8564209f
--- /dev/null
+++ b/examples/03_variogram/00_fit_variogram.py
@@ -0,0 +1,34 @@
+"""
+Fit Variogram
+-------------
+"""
+import numpy as np
+import gstools as gs
+
+###############################################################################
+# Generate a synthetic field with an exponential model.
+
+x = np.random.RandomState(19970221).rand(1000) * 100.0
+y = np.random.RandomState(20011012).rand(1000) * 100.0
+model = gs.Exponential(dim=2, var=2, len_scale=8)
+srf = gs.SRF(model, mean=0, seed=19970221)
+field = srf((x, y))
+
+###############################################################################
+# Estimate the variogram of the field with 40 bins.
+
+bins = np.arange(40)
+bin_center, gamma = gs.vario_estimate_unstructured((x, y), field, bins)
+
+###############################################################################
+# Fit the variogram with a stable model (no nugget fitted).
+
+fit_model = gs.Stable(dim=2)
+fit_model.fit_variogram(bin_center, gamma, nugget=False)
+
+###############################################################################
+# Plot the fitting result.
+
+ax = fit_model.plot(x_max=40)
+ax.plot(bin_center, gamma)
+print(fit_model)
diff --git a/examples/03_variogram/01_variogram_estimation.py b/examples/03_variogram/01_variogram_estimation.py
new file mode 100644
index 00000000..6685eb40
--- /dev/null
+++ b/examples/03_variogram/01_variogram_estimation.py
@@ -0,0 +1,286 @@
+"""
+An Example with Actual Data
+---------------------------
+
+This example is going to be a bit more extensive and we are going to do some
+basic data preprocessing for the actual variogram estimation. But this example
+will be self-contained and all data gathering and processing will be done in
+this example script.
+
+
+The Data
+^^^^^^^^
+
+We are going to analyse the Herten aquifer, which is situated in Southern
+Germany. Multiple outcrop faces where surveyed and interpolated to a 3D
+dataset. In these publications, you can find more information about the data:
+
+| Bayer, Peter; Comunian, Alessandro; Höyng, Dominik; Mariethoz, Gregoire (2015): Physicochemical properties and 3D geostatistical simulations of the Herten and the Descalvado aquifer analogs. PANGAEA, https://doi.org/10.1594/PANGAEA.844167,
+| Supplement to: Bayer, P et al. (2015): Three-dimensional multi-facies realizations of sedimentary reservoir and aquifer analogs. Scientific Data, 2, 150033, https://doi.org/10.1038/sdata.2015.33
+|
+
+Retrieving the Data
+^^^^^^^^^^^^^^^^^^^
+
+To begin with, we need to download and extract the data. Therefore, we are
+going to use some built-in Python libraries. For simplicity, many values and
+strings will be hardcoded.
+
+You don't have to execute the ``download_herten`` and ``generate_transmissivity``
+functions, since the only produce the ``herten_transmissivity.gz``
+and ``grid_dim_origin_spacing.txt``, which are already present.
+"""
+import os
+import numpy as np
+import matplotlib.pyplot as plt
+import gstools as gs
+
+VTK_PATH = os.path.join("Herten-analog", "sim-big_1000x1000x140", "sim.vtk")
+
+###############################################################################
+
+
+def download_herten():
+ """Download the data, warning: its about 250MB."""
+ import zipfile
+ import urllib.request
+
+ print("Downloading Herten data")
+ data_filename = "data.zip"
+ data_url = (
+ "http://store.pangaea.de/Publications/"
+ "Bayer_et_al_2015/Herten-analog.zip"
+ )
+ urllib.request.urlretrieve(data_url, "data.zip")
+ # extract the "big" simulation
+ with zipfile.ZipFile(data_filename, "r") as zf:
+ zf.extract(VTK_PATH)
+
+
+###############################################################################
+
+
+def generate_transmissivity():
+ """Generate a file with a transmissivity field from the HERTEN data."""
+ import pyvista as pv
+ import shutil
+
+ print("Loading Herten data with pyvista")
+ mesh = pv.read(VTK_PATH)
+ herten = mesh.point_arrays["facies"].reshape(mesh.dimensions, order="F")
+ # conductivity values per fazies from the supplementary data
+ cond = 1e-4 * np.array(
+ [2.5, 2.3, 0.61, 260, 1300, 950, 0.43, 0.006, 23, 1.4]
+ )
+ # asign the conductivities to the facies
+ herten_cond = cond[herten]
+ # Next, we are going to calculate the transmissivity,
+ # by integrating over the vertical axis
+ herten_trans = np.sum(herten_cond, axis=2) * mesh.spacing[2]
+ # saving some grid informations
+ grid = [mesh.dimensions[:2], mesh.origin[:2], mesh.spacing[:2]]
+ print("Saving the transmissivity field and grid information")
+ np.savetxt("herten_transmissivity.gz", herten_trans)
+ np.savetxt("grid_dim_origin_spacing.txt", grid)
+ # Some cleanup. You can comment out these lines to keep the downloaded data
+ os.remove("data.zip")
+ shutil.rmtree("Herten-analog")
+
+
+###############################################################################
+# Downloading and Preprocessing
+# ^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
+#
+# You can uncomment the following two calls, so the data is downloaded
+# and processed again.
+
+# download_herten()
+# generate_transmissivity()
+
+
+###############################################################################
+# Analyzing the data
+# ^^^^^^^^^^^^^^^^^^
+#
+# The Herten data provides information about the grid, which was already used in
+# the previous code block. From this information, we can create our own grid on
+# which we can estimate the variogram. As a first step, we are going to estimate
+# an isotropic variogram, meaning that we will take point pairs from all
+# directions into account. An unstructured grid is a natural choice for this.
+# Therefore, we are going to create an unstructured grid from the given,
+# structured one. For this, we are going to write another small function
+
+herten_log_trans = np.log(np.loadtxt("herten_transmissivity.gz"))
+dim, origin, spacing = np.loadtxt("grid_dim_origin_spacing.txt")
+
+# create a structured grid on which the data is defined
+x_s = np.arange(origin[0], origin[0] + dim[0] * spacing[0], spacing[0])
+y_s = np.arange(origin[1], origin[1] + dim[1] * spacing[1], spacing[1])
+# create the corresponding unstructured grid for the variogram estimation
+x_u, y_u = np.meshgrid(x_s, y_s)
+
+
+###############################################################################
+# Let's have a look at the transmissivity field of the Herten aquifer
+
+plt.imshow(herten_log_trans.T, origin="lower", aspect="equal")
+plt.show()
+
+
+###############################################################################
+# Estimating the Variogram
+# ^^^^^^^^^^^^^^^^^^^^^^^^
+#
+# Finally, everything is ready for the variogram estimation. For the unstructured
+# method, we have to define the bins on which the variogram will be estimated.
+# Through expert knowledge (i.e. fiddling around), we assume that the main
+# features of the variogram will be below 10 metres distance. And because the
+# data has a high spatial resolution, the resolution of the bins can also be
+# high. The transmissivity data is still defined on a structured grid, but we can
+# simply flatten it with :any:`numpy.ndarray.flatten`, in order to bring it into
+# the right shape. It might be more memory efficient to use
+# ``herten_log_trans.reshape(-1)``, but for better readability, we will stick to
+# :any:`numpy.ndarray.flatten`. Taking all data points into account would take a
+# very long time (expert knowledge \*wink\*), thus we will only take 2000 datapoints into account, which are sampled randomly. In order to make the exact
+# results reproducible, we can also set a seed.
+
+
+bins = np.linspace(0, 10, 50)
+bin_center, gamma = gs.vario_estimate_unstructured(
+ (x_u, y_u),
+ herten_log_trans.reshape(-1),
+ bins,
+ sampling_size=2000,
+ sampling_seed=19920516,
+)
+
+###############################################################################
+# The estimated variogram is calculated on the centre of the given bins,
+# therefore, the ``bin_center`` array is also returned.
+
+###############################################################################
+# Fitting the Variogram
+# ^^^^^^^^^^^^^^^^^^^^^
+#
+# Now, we can see, if the estimated variogram can be modelled by a common
+# variogram model. Let's try the :any:`Exponential` model.
+
+# fit an exponential model
+fit_model = gs.Exponential(dim=2)
+fit_model.fit_variogram(bin_center, gamma, nugget=False)
+
+###############################################################################
+# Finally, we can visualise some results. For quickly plotting a covariance
+# model, GSTools provides some helper functions.
+
+ax = fit_model.plot(x_max=max(bin_center))
+ax.plot(bin_center, gamma)
+
+
+###############################################################################
+# That looks like a pretty good fit! By printing the model, we can directly see
+# the fitted parameters
+
+print(fit_model)
+
+###############################################################################
+# With this data, we could start generating new ensembles of the Herten aquifer
+# with the :any:`SRF` class.
+
+
+###############################################################################
+# Estimating the Variogram in Specific Directions
+# ^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
+#
+# Estimating a variogram on a structured grid gives us the possibility to only
+# consider values in a specific direction. This could be a first test, to see if
+# the data is anisotropic.
+# In order to speed up the calculations, we are going to only use every 10th datapoint and for a comparison with the isotropic variogram calculated earlier, we
+# only need the first 21 array items.
+
+
+# estimate the variogram on a structured grid
+# use only every 10th value, otherwise calculations would take very long
+x_s_skip = np.ravel(x_s)[::10]
+y_s_skip = np.ravel(y_s)[::10]
+herten_trans_skip = herten_log_trans[::10, ::10]
+
+###############################################################################
+# With this much smaller data set, we can immediately estimate the variogram in
+# the x- and y-axis
+
+gamma_x = gs.vario_estimate_structured(herten_trans_skip, direction="x")
+gamma_y = gs.vario_estimate_structured(herten_trans_skip, direction="y")
+
+###############################################################################
+# With these two estimated variograms, we can start fitting :any:`Exponential`
+# covariance models
+
+x_plot = x_s_skip[:21]
+y_plot = y_s_skip[:21]
+# fit an exponential model
+fit_model_x = gs.Exponential(dim=2)
+fit_model_x.fit_variogram(x_plot, gamma_x[:21], nugget=False)
+fit_model_y = gs.Exponential(dim=2)
+fit_model_y.fit_variogram(y_plot, gamma_y[:21], nugget=False)
+
+###############################################################################
+# Now, the isotropic variogram and the two variograms in x- and y-direction can
+# be plotted together with their respective models, which will be plotted with
+# dashed lines.
+
+plt.figure() # new figure
+line, = plt.plot(bin_center, gamma, label="estimated variogram (isotropic)")
+plt.plot(
+ bin_center,
+ fit_model.variogram(bin_center),
+ color=line.get_color(),
+ linestyle="--",
+ label="exp. variogram (isotropic)",
+)
+
+line, = plt.plot(x_plot, gamma_x[:21], label="estimated variogram in x-dir")
+plt.plot(
+ x_plot,
+ fit_model_x.variogram(x_plot),
+ color=line.get_color(),
+ linestyle="--",
+ label="exp. variogram in x-dir",
+)
+
+line, = plt.plot(y_plot, gamma_y[:21], label="estimated variogram in y-dir")
+plt.plot(
+ y_plot,
+ fit_model_y.variogram(y_plot),
+ color=line.get_color(),
+ linestyle="--",
+ label="exp. variogram in y-dir",
+)
+
+plt.legend()
+plt.show()
+
+###############################################################################
+# The plot might be a bit cluttered, but at least it is pretty obvious that the
+# Herten aquifer has no apparent anisotropies in its spatial structure.
+
+print("semivariogram model (isotropic):\n", fit_model)
+print("semivariogram model (in x-dir.):\n", fit_model_x)
+print("semivariogram model (in y-dir.):\n", fit_model_y)
+
+
+###############################################################################
+# Creating a Spatial Random Field from the Herten Parameters
+# ^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^^
+#
+# With all the hard work done, it's straight forward now, to generate new
+# *Herten-like realisations*
+
+# create a spatial random field on the low-resolution grid
+srf = gs.SRF(fit_model, seed=19770928)
+srf.structured([x_s_skip, y_s_skip])
+ax = srf.plot()
+ax.set_aspect("equal")
+
+###############################################################################
+# That's pretty neat!
diff --git a/examples/03_variogram/README.rst b/examples/03_variogram/README.rst
new file mode 100644
index 00000000..d16c16c2
--- /dev/null
+++ b/examples/03_variogram/README.rst
@@ -0,0 +1,14 @@
+Tutorial 3: Variogram Estimation
+================================
+
+Estimating the spatial correlations is an important part of geostatistics.
+These spatial correlations can be expressed by the variogram, which can be
+estimated with the subpackage :any:`gstools.variogram`. The variograms can be
+estimated on structured and unstructured grids.
+
+The same `(semi-)variogram `_ as
+:ref:`tutorial_02_cov` is being used
+by this subpackage.
+
+Gallery
+-------
diff --git a/examples/03_variogram/grid_dim_origin_spacing.txt b/examples/03_variogram/grid_dim_origin_spacing.txt
new file mode 100644
index 00000000..024928d3
--- /dev/null
+++ b/examples/03_variogram/grid_dim_origin_spacing.txt
@@ -0,0 +1,3 @@
+1.000000000000000000e+03 1.000000000000000000e+03
+0.000000000000000000e+00 0.000000000000000000e+00
+5.000000000000000278e-02 5.000000000000000278e-02
diff --git a/examples/03_variogram/herten_transmissivity.gz b/examples/03_variogram/herten_transmissivity.gz
new file mode 100644
index 00000000..fa1d00e3
Binary files /dev/null and b/examples/03_variogram/herten_transmissivity.gz differ
diff --git a/examples/04_export.py b/examples/04_export.py
deleted file mode 100644
index bb9cb439..00000000
--- a/examples/04_export.py
+++ /dev/null
@@ -1,7 +0,0 @@
-from gstools import SRF, Gaussian
-
-x = y = range(100)
-model = Gaussian(dim=2, var=1, len_scale=10)
-srf = SRF(model)
-field = srf((x, y), mesh_type="structured")
-srf.vtk_export("field")
diff --git a/examples/04_vector_field/00_vector_field.py b/examples/04_vector_field/00_vector_field.py
new file mode 100644
index 00000000..e2c05ee9
--- /dev/null
+++ b/examples/04_vector_field/00_vector_field.py
@@ -0,0 +1,45 @@
+"""
+Generating a Random Vector Field
+--------------------------------
+
+As a first example we are going to generate a vector field with a Gaussian
+covariance model on a structured grid:
+"""
+import numpy as np
+import gstools as gs
+
+# the grid
+x = np.arange(100)
+y = np.arange(100)
+
+# a smooth Gaussian covariance model
+model = gs.Gaussian(dim=2, var=1, len_scale=10)
+srf = gs.SRF(model, generator="VectorField", seed=19841203)
+srf((x, y), mesh_type="structured")
+srf.plot()
+
+###############################################################################
+# Let us have a look at the influence of the covariance model. Choosing the
+# exponential model and keeping all other parameters the same
+
+# a rougher exponential covariance model
+model2 = gs.Exponential(dim=2, var=1, len_scale=10)
+srf.model = model2
+srf((x, y), mesh_type="structured", seed=19841203)
+srf.plot()
+
+###############################################################################
+# and we see, that the wiggles are much "rougher" than the smooth Gaussian ones.
+
+
+###############################################################################
+# Applications
+# ------------
+#
+# One great advantage of the Kraichnan method is, that after some initializations,
+# one can compute the velocity field at arbitrary points, online, with hardly any
+# overhead.
+# This means, that for a Lagrangian transport simulation for example, the velocity
+# can be evaluated at each particle position very efficiently and without any
+# interpolation. These field interpolations are a common problem for Lagrangian
+# methods.
diff --git a/examples/04_vector_field/README.rst b/examples/04_vector_field/README.rst
new file mode 100644
index 00000000..1875531e
--- /dev/null
+++ b/examples/04_vector_field/README.rst
@@ -0,0 +1,36 @@
+Tutorial 4: Random Vector Field Generation
+==========================================
+
+In 1970, Kraichnan was the first to suggest a randomization method.
+For studying the diffusion of single particles in a random incompressible
+velocity field, he came up with a randomization method which includes a
+projector which ensures the incompressibility of the vector field.
+
+
+Without loss of generality we assume that the mean velocity :math:`\bar{U}` is oriented
+towards the direction of the first basis vector :math:`\mathbf{e}_1`. Our goal is now to
+generate random fluctuations with a given covariance model around this mean velocity.
+And at the same time, making sure that the velocity field remains incompressible or
+in other words, ensure :math:`\nabla \cdot \mathbf U = 0`.
+This can be done by using the randomization method we already know, but adding a
+projector to every mode being summed:
+
+
+.. math::
+
+ \mathbf{U}(\mathbf{x}) = \bar{U} \mathbf{e}_1 - \sqrt{\frac{\sigma^{2}}{N}}
+ \sum_{i=1}^{N} \mathbf{p}(\mathbf{k}_i) \left[ Z_{1,i}
+ \cos\left( \langle \mathbf{k}_{i}, \mathbf{x} \rangle \right)
+ + \sin\left( \langle \mathbf{k}_{i}, \mathbf{x} \rangle \right) \right]
+
+with the projector
+
+.. math::
+
+ \mathbf{p}(\mathbf{k}_i) = \mathbf{e}_1 - \frac{\mathbf{k}_i k_1}{k^2} \; .
+
+By calculating :math:`\nabla \cdot \mathbf U = 0`, it can be verified, that
+the resulting field is indeed incompressible.
+
+Gallery
+-------
diff --git a/examples/05_kriging/00_simple_kriging.py b/examples/05_kriging/00_simple_kriging.py
new file mode 100755
index 00000000..c7bcdcb2
--- /dev/null
+++ b/examples/05_kriging/00_simple_kriging.py
@@ -0,0 +1,48 @@
+r"""
+Simple Kriging
+--------------
+
+Simple kriging assumes a known mean of the data.
+For simplicity we assume a mean of 0,
+which can be achieved by subtracting the mean from the observed values and
+subsequently adding it to the resulting data.
+
+The resulting equation system for :math:`W` is given by:
+
+.. math::
+
+ W = \begin{pmatrix}c(x_1,x_1) & \cdots & c(x_1,x_n) \\
+ \vdots & \ddots & \vdots \\
+ c(x_n,x_1) & \cdots & c(x_n,x_n)
+ \end{pmatrix}^{-1}
+ \begin{pmatrix}c(x_1,x_0) \\ \vdots \\ c(x_n,x_0) \end{pmatrix}
+
+Thereby :math:`c(x_i,x_j)` is the covariance of the given observations.
+
+
+Example
+^^^^^^^
+
+Here we use simple kriging in 1D (for plotting reasons) with 5 given observations/conditions.
+The mean of the field has to be given beforehand.
+
+"""
+import numpy as np
+from gstools import Gaussian, krige
+
+# condtions
+cond_pos = [0.3, 1.9, 1.1, 3.3, 4.7]
+cond_val = [0.47, 0.56, 0.74, 1.47, 1.74]
+# resulting grid
+gridx = np.linspace(0.0, 15.0, 151)
+# spatial random field class
+model = Gaussian(dim=1, var=0.5, len_scale=2)
+
+###############################################################################
+krig = krige.Simple(model, mean=1, cond_pos=cond_pos, cond_val=cond_val)
+krig(gridx)
+
+###############################################################################
+ax = krig.plot()
+ax.scatter(cond_pos, cond_val, color="k", zorder=10, label="Conditions")
+ax.legend()
diff --git a/examples/05_kriging/01_ordinary_kriging.py b/examples/05_kriging/01_ordinary_kriging.py
new file mode 100644
index 00000000..2e56c797
--- /dev/null
+++ b/examples/05_kriging/01_ordinary_kriging.py
@@ -0,0 +1,48 @@
+r"""
+Ordinary Kriging
+----------------
+
+Ordinary kriging will estimate an appropriate mean of the field,
+based on the given observations/conditions and the covariance model used.
+
+The resulting system of equations for :math:`W` is given by:
+
+.. math::
+
+ \begin{pmatrix}W\\\mu\end{pmatrix} = \begin{pmatrix}
+ \gamma(x_1,x_1) & \cdots & \gamma(x_1,x_n) &1 \\
+ \vdots & \ddots & \vdots & \vdots \\
+ \gamma(x_n,x_1) & \cdots & \gamma(x_n,x_n) & 1 \\
+ 1 &\cdots& 1 & 0
+ \end{pmatrix}^{-1}
+ \begin{pmatrix}\gamma(x_1,x_0) \\ \vdots \\ \gamma(x_n,x_0) \\ 1\end{pmatrix}
+
+Thereby :math:`\gamma(x_i,x_j)` is the semi-variogram of the given observations
+and :math:`\mu` is a Lagrange multiplier to minimize the kriging error and estimate the mean.
+
+
+Example
+^^^^^^^
+
+Here we use ordinary kriging in 1D (for plotting reasons) with 5 given observations/conditions.
+The estimated mean can be accessed by ``krig.mean``.
+"""
+import numpy as np
+from gstools import Gaussian, krige
+
+# condtions
+cond_pos = [0.3, 1.9, 1.1, 3.3, 4.7]
+cond_val = [0.47, 0.56, 0.74, 1.47, 1.74]
+# resulting grid
+gridx = np.linspace(0.0, 15.0, 151)
+# spatial random field class
+model = Gaussian(dim=1, var=0.5, len_scale=2)
+
+###############################################################################
+krig = krige.Ordinary(model, cond_pos=cond_pos, cond_val=cond_val)
+krig(gridx)
+
+###############################################################################
+ax = krig.plot()
+ax.scatter(cond_pos, cond_val, color="k", zorder=10, label="Conditions")
+ax.legend()
diff --git a/examples/05_kriging/02_pykrige_interface.py b/examples/05_kriging/02_pykrige_interface.py
new file mode 100755
index 00000000..724ff06c
--- /dev/null
+++ b/examples/05_kriging/02_pykrige_interface.py
@@ -0,0 +1,63 @@
+"""
+Interface to PyKrige
+--------------------
+
+To use fancier methods like
+`regression kriging `__,
+we provide an interface to
+`PyKrige `__.
+
+In the future you can pass a GSTools Covariance Model
+to the PyKrige routines as ``variogram_model``.
+
+At the moment we only provide prepared
+keyword arguments for the pykrige routines.
+
+To demonstrate the general workflow, we compare the ordinary kriging of PyKrige
+with GSTools in 2D:
+"""
+import numpy as np
+import gstools as gs
+from pykrige.ok import OrdinaryKriging
+from matplotlib import pyplot as plt
+
+# conditioning data
+data = np.array(
+ [
+ [0.3, 1.2, 0.47],
+ [1.9, 0.6, 0.56],
+ [1.1, 3.2, 0.74],
+ [3.3, 4.4, 1.47],
+ [4.7, 3.8, 1.74],
+ ]
+)
+
+# grid definition for output field
+gridx = np.arange(0.0, 5.5, 0.1)
+gridy = np.arange(0.0, 6.5, 0.1)
+
+###############################################################################
+# A GSTools based covariance model.
+
+cov_model = gs.Gaussian(
+ dim=2, len_scale=1, anis=0.2, angles=-0.5, var=0.5, nugget=0.1
+)
+
+###############################################################################
+# Ordinary kriging with pykrige.
+# A dictionary containing keyword arguments for the pykrige routines is
+# provided by the gstools covariance models.
+
+pk_kwargs = cov_model.pykrige_kwargs
+OK1 = OrdinaryKriging(data[:, 0], data[:, 1], data[:, 2], **pk_kwargs)
+z1, ss1 = OK1.execute("grid", gridx, gridy)
+plt.imshow(z1, origin="lower")
+plt.show()
+
+###############################################################################
+# Ordinary kriging with gstools for comparison.
+
+OK2 = gs.krige.Ordinary(cov_model, [data[:, 0], data[:, 1]], data[:, 2])
+OK2.structured([gridx, gridy])
+ax = OK2.plot()
+ax.set_aspect("equal")
diff --git a/examples/12_compare_kriging.py b/examples/05_kriging/03_compare_kriging.py
similarity index 63%
rename from examples/12_compare_kriging.py
rename to examples/05_kriging/03_compare_kriging.py
index b22f05ee..4b9925f5 100755
--- a/examples/12_compare_kriging.py
+++ b/examples/05_kriging/03_compare_kriging.py
@@ -1,3 +1,7 @@
+"""
+Compare Kriging
+---------------
+"""
import numpy as np
from gstools import Gaussian, krige
import matplotlib.pyplot as plt
@@ -7,12 +11,22 @@
cond_val = [0.47, 0.56, 0.74, 1.47, 1.74]
# resulting grid
gridx = np.linspace(0.0, 15.0, 151)
-# spatial random field class
+
+###############################################################################
+# A gaussian variogram model.
+
model = Gaussian(dim=1, var=0.5, len_scale=2)
+
+###############################################################################
+# Two kriged fields. One with simple and one with ordinary kriging.
+
kr1 = krige.Simple(model=model, mean=1, cond_pos=cond_pos, cond_val=cond_val)
kr2 = krige.Ordinary(model=model, cond_pos=cond_pos, cond_val=cond_val)
kr1(gridx)
kr2(gridx)
+
+###############################################################################
+
plt.plot(gridx, kr1.field, label="simple kriged field")
plt.plot(gridx, kr2.field, label="ordinary kriged field")
plt.scatter(cond_pos, cond_val, color="k", zorder=10, label="Conditions")
diff --git a/examples/05_kriging/04_extdrift_kriging.py b/examples/05_kriging/04_extdrift_kriging.py
new file mode 100755
index 00000000..d7cc8e29
--- /dev/null
+++ b/examples/05_kriging/04_extdrift_kriging.py
@@ -0,0 +1,24 @@
+"""
+External Drift Kriging
+----------------------
+"""
+import numpy as np
+from gstools import SRF, Gaussian, krige
+
+# synthetic condtions with a drift
+drift_model = Gaussian(dim=1, len_scale=4)
+drift = SRF(drift_model, seed=1010)
+cond_pos = [0.3, 1.9, 1.1, 3.3, 4.7]
+ext_drift = drift(cond_pos)
+cond_val = ext_drift * 2 + 1
+# resulting grid
+gridx = np.linspace(0.0, 15.0, 151)
+grid_drift = drift(gridx)
+# kriging
+model = Gaussian(dim=1, var=2, len_scale=4)
+krig = krige.ExtDrift(model, cond_pos, cond_val, ext_drift)
+krig(gridx, ext_drift=grid_drift)
+ax = krig.plot()
+ax.scatter(cond_pos, cond_val, color="k", zorder=10, label="Conditions")
+ax.plot(gridx, grid_drift, label="drift")
+ax.legend()
diff --git a/examples/05_kriging/05_universal_kriging.py b/examples/05_kriging/05_universal_kriging.py
new file mode 100755
index 00000000..cf93da0c
--- /dev/null
+++ b/examples/05_kriging/05_universal_kriging.py
@@ -0,0 +1,24 @@
+"""
+Universal Kriging
+-----------------
+"""
+import numpy as np
+from gstools import SRF, Gaussian, krige
+
+# synthetic condtions with a drift
+drift_model = Gaussian(dim=1, var=0.1, len_scale=2)
+drift = SRF(drift_model, seed=101)
+cond_pos = np.linspace(0.1, 8, 10)
+cond_val = drift(cond_pos) + cond_pos * 0.1 + 1
+# resulting grid
+gridx = np.linspace(0.0, 15.0, 151)
+drift_field = drift(gridx) + gridx * 0.1 + 1
+# kriging
+model = Gaussian(dim=1, var=0.1, len_scale=2)
+krig = krige.Universal(model, cond_pos, cond_val, "linear")
+krig(gridx)
+ax = krig.plot()
+ax.scatter(cond_pos, cond_val, color="k", zorder=10, label="Conditions")
+ax.plot(gridx, gridx * 0.1 + 1, ":", label="linear drift")
+ax.plot(gridx, drift_field, "--", label="original field")
+ax.legend()
diff --git a/examples/05_kriging/06_detrended_kriging.py b/examples/05_kriging/06_detrended_kriging.py
new file mode 100755
index 00000000..9d9fbc06
--- /dev/null
+++ b/examples/05_kriging/06_detrended_kriging.py
@@ -0,0 +1,30 @@
+"""
+Detrended Kriging
+-----------------
+"""
+import numpy as np
+from gstools import SRF, Gaussian, krige
+
+
+def trend(x):
+ """Example for a simple linear trend."""
+ return x * 0.1 + 1
+
+
+# synthetic condtions with trend/drift
+drift_model = Gaussian(dim=1, var=0.1, len_scale=2)
+drift = SRF(drift_model, seed=101)
+cond_pos = np.linspace(0.1, 8, 10)
+cond_val = drift(cond_pos) + trend(cond_pos)
+# resulting grid
+gridx = np.linspace(0.0, 15.0, 151)
+drift_field = drift(gridx) + trend(gridx)
+# kriging
+model = Gaussian(dim=1, var=0.1, len_scale=2)
+krig_trend = krige.Detrended(model, cond_pos, cond_val, trend)
+krig_trend(gridx)
+ax = krig_trend.plot()
+ax.scatter(cond_pos, cond_val, color="k", zorder=10, label="Conditions")
+ax.plot(gridx, trend(gridx), ":", label="linear trend")
+ax.plot(gridx, drift_field, "--", label="original field")
+ax.legend()
diff --git a/examples/05_kriging/07_detrended_ordinary_kriging.py b/examples/05_kriging/07_detrended_ordinary_kriging.py
new file mode 100755
index 00000000..585fdcd1
--- /dev/null
+++ b/examples/05_kriging/07_detrended_ordinary_kriging.py
@@ -0,0 +1,30 @@
+"""
+Detrended Ordinary Kriging
+--------------------------
+"""
+import numpy as np
+from gstools import SRF, Gaussian, krige
+
+
+def trend(x):
+ """Example for a simple linear trend."""
+ return x * 0.1 + 1
+
+
+# synthetic condtions with trend/drift
+drift_model = Gaussian(dim=1, var=0.1, len_scale=2)
+drift = SRF(drift_model, seed=101)
+cond_pos = np.linspace(0.1, 8, 10)
+cond_val = drift(cond_pos) + trend(cond_pos)
+# resulting grid
+gridx = np.linspace(0.0, 15.0, 151)
+drift_field = drift(gridx) + trend(gridx)
+# kriging
+model = Gaussian(dim=1, var=0.1, len_scale=2)
+krig_trend = krige.Ordinary(model, cond_pos, cond_val, trend)
+krig_trend(gridx)
+ax = krig_trend.plot()
+ax.scatter(cond_pos, cond_val, color="k", zorder=10, label="Conditions")
+ax.plot(gridx, trend(gridx), ":", label="linear trend")
+ax.plot(gridx, drift_field, "--", label="original field")
+ax.legend()
diff --git a/examples/05_kriging/README.rst b/examples/05_kriging/README.rst
new file mode 100644
index 00000000..f0d9ef71
--- /dev/null
+++ b/examples/05_kriging/README.rst
@@ -0,0 +1,57 @@
+.. _tutorial_05_kriging:
+
+Tutorial 5: Kriging
+===================
+
+The subpackage :py:mod:`gstools.krige` provides routines for Gaussian process regression, also known as kriging.
+Kriging is a method of data interpolation based on predefined covariance models.
+
+The aim of kriging is to derive the value of a field at some point :math:`x_0`,
+when there are fixed observed values :math:`z(x_1)\ldots z(x_n)` at given points :math:`x_i`.
+
+The resluting value :math:`z_0` at :math:`x_0` is calculated as a weighted mean:
+
+.. math::
+
+ z_0 = \sum_{i=1}^n w_i \cdot z_i
+
+The weights :math:`W = (w_1,\ldots,w_n)` depent on the given covariance model and the location of the target point.
+
+The different kriging approaches provide different ways of calculating :math:`W`.
+
+
+The routines for kriging are almost identical to the routines for spatial random fields.
+First you define a covariance model, as described in :ref:`tutorial_02_cov`,
+then you initialize the kriging class with this model:
+
+.. code-block:: python
+
+ import gstools as gs
+ # condtions
+ cond_pos = [...]
+ cond_val = [...]
+ model = gs.Gaussian(dim=1, var=0.5, len_scale=2)
+ krig = gs.krige.Simple(model, cond_pos=cond_pos, cond_val=cond_val, mean=1)
+
+The resulting field instance ``krig`` has the same methods as the
+:any:`SRF` class.
+You can call it to evaluate the kriged field at different points,
+you can plot the latest field or you can export the field and so on.
+
+Provided Kriging Methods
+------------------------
+
+.. currentmodule:: gstools.krige
+
+The following kriging methods are provided within the
+submodule :any:`gstools.krige`.
+
+.. autosummary::
+ Simple
+ Ordinary
+ Universal
+ ExtDrift
+ Detrended
+
+Gallery
+-------
diff --git a/examples/05_srf_ensemble.py b/examples/05_srf_ensemble.py
deleted file mode 100644
index 3db77e0a..00000000
--- a/examples/05_srf_ensemble.py
+++ /dev/null
@@ -1,22 +0,0 @@
-import numpy as np
-import matplotlib.pyplot as pt
-from gstools import SRF, Gaussian
-from gstools.random import MasterRNG
-
-x = y = np.arange(100)
-
-model = Gaussian(dim=2, var=1, len_scale=10)
-srf = SRF(model)
-
-ens_no = 4
-field = []
-seed = MasterRNG(20170519)
-for i in range(ens_no):
- field.append(srf((x, y), seed=seed(), mesh_type="structured"))
-
-fig, ax = pt.subplots(2, 2, sharex=True, sharey=True)
-ax = ax.flatten()
-for i in range(ens_no):
- ax[i].imshow(field[i].T, origin="lower")
-
-pt.show()
diff --git a/examples/06_conditioned_fields/00_condition_ensemble.py b/examples/06_conditioned_fields/00_condition_ensemble.py
new file mode 100644
index 00000000..ca9ab20f
--- /dev/null
+++ b/examples/06_conditioned_fields/00_condition_ensemble.py
@@ -0,0 +1,42 @@
+"""
+Example: Conditioning with Ordinary Kriging
+-------------------------------------------
+
+Here we use ordinary kriging in 1D (for plotting reasons) with 5 given observations/conditions,
+to generate an ensemble of conditioned random fields.
+The estimated mean can be accessed by ``srf.mean``.
+"""
+import numpy as np
+import matplotlib.pyplot as plt
+import gstools as gs
+
+# condtions
+cond_pos = [0.3, 1.9, 1.1, 3.3, 4.7]
+cond_val = [0.47, 0.56, 0.74, 1.47, 1.74]
+gridx = np.linspace(0.0, 15.0, 151)
+
+###############################################################################
+
+# spatial random field class
+model = gs.Gaussian(dim=1, var=0.5, len_scale=2)
+srf = gs.SRF(model)
+srf.set_condition(cond_pos, cond_val, "ordinary")
+
+###############################################################################
+
+fields = []
+for i in range(100):
+ # print(i) if i % 10 == 0 else None
+ fields.append(srf(gridx, seed=i))
+ label = "Conditioned ensemble" if i == 0 else None
+ plt.plot(gridx, fields[i], color="k", alpha=0.1, label=label)
+plt.plot(gridx, np.full_like(gridx, srf.mean), label="estimated mean")
+plt.plot(gridx, np.mean(fields, axis=0), linestyle=":", label="Ensemble mean")
+plt.plot(gridx, srf.krige_field, linestyle="dashed", label="kriged field")
+plt.scatter(cond_pos, cond_val, color="k", zorder=10, label="Conditions")
+plt.legend()
+plt.show()
+
+###############################################################################
+# As you can see, the kriging field coincides with the ensemble mean of the
+# conditioned random fields and the estimated mean is the mean of the far-field.
diff --git a/examples/06_conditioned_fields/README.rst b/examples/06_conditioned_fields/README.rst
new file mode 100644
index 00000000..bd5a6d9d
--- /dev/null
+++ b/examples/06_conditioned_fields/README.rst
@@ -0,0 +1,34 @@
+Tutorial 6: Conditioned Fields
+==============================
+
+Kriged fields tend to approach the field mean outside the area of observations.
+To generate random fields, that coincide with given observations, but are still
+random according to a given covariance model away from the observations proximity,
+we provide the generation of conditioned random fields.
+
+
+The idea behind conditioned random fields builds up on kriging.
+First we generate a field with a kriging method, then we generate a random field,
+and finally we generate another kriged field to eliminate the error between
+the random field and the kriged field of the given observations.
+
+To do so, you can choose between ordinary and simple kriging.
+In case of ordinary kriging, the mean of the SRF will be overwritten by the
+estimated mean.
+
+The setup of the spatial random field is the same as described in
+:ref:`tutorial_02_cov`.
+You just need to add the conditions as described in :ref:`tutorial_05_kriging`:
+
+.. code-block:: python
+
+ srf.set_condition(cond_pos, cond_val, "simple")
+
+or:
+
+.. code-block:: python
+
+ srf.set_condition(cond_pos, cond_val, "ordinary")
+
+Gallery
+-------
diff --git a/examples/06_unstr_srf_export.py b/examples/06_unstr_srf_export.py
deleted file mode 100644
index 8ea844ca..00000000
--- a/examples/06_unstr_srf_export.py
+++ /dev/null
@@ -1,23 +0,0 @@
-import numpy as np
-import matplotlib.pyplot as pt
-from gstools import SRF, Exponential
-from gstools.random import MasterRNG
-
-# creating our own unstructured grid
-seed = MasterRNG(19970221)
-rng = np.random.RandomState(seed())
-x = rng.randint(0, 100, size=10000)
-y = rng.randint(0, 100, size=10000)
-
-model = Exponential(dim=2, var=1, len_scale=[12.0, 3.0], angles=np.pi / 8.0)
-
-srf = SRF(model, seed=20170519)
-
-field = srf((x, y))
-srf.vtk_export("field")
-# Or create a PyVista dataset
-# mesh = srf.to_pyvista()
-
-pt.tricontourf(x, y, field.T)
-pt.axes().set_aspect("equal")
-pt.show()
diff --git a/examples/07_srf_merge.py b/examples/07_srf_merge.py
deleted file mode 100644
index 3f3cdb3e..00000000
--- a/examples/07_srf_merge.py
+++ /dev/null
@@ -1,29 +0,0 @@
-import numpy as np
-import matplotlib.pyplot as pt
-from gstools import SRF, Exponential
-from gstools.random import MasterRNG
-
-# creating our own unstructured grid
-seed = MasterRNG(19970221)
-rng = np.random.RandomState(seed())
-x = rng.randint(0, 100, size=10000)
-y = rng.randint(0, 100, size=10000)
-
-model = Exponential(dim=2, var=1, len_scale=[12.0, 3.0], angles=np.pi / 8.0)
-
-srf = SRF(model, seed=20170519)
-
-field = srf((x, y))
-
-# new grid
-seed = MasterRNG(20011012)
-rng = np.random.RandomState(seed())
-x2 = rng.randint(99, 150, size=10000)
-y2 = rng.randint(20, 80, size=10000)
-
-field2 = srf((x2, y2))
-
-pt.tricontourf(x, y, field.T)
-pt.tricontourf(x2, y2, field2.T)
-pt.axes().set_aspect("equal")
-pt.show()
diff --git a/examples/07_transformations/00_log_normal.py b/examples/07_transformations/00_log_normal.py
new file mode 100755
index 00000000..a44f50fb
--- /dev/null
+++ b/examples/07_transformations/00_log_normal.py
@@ -0,0 +1,15 @@
+"""
+log-normal fields
+-----------------
+
+Here we transform a field to a log-normal distribution:
+"""
+import gstools as gs
+
+# structured field with a size of 100x100 and a grid-size of 1x1
+x = y = range(100)
+model = gs.Gaussian(dim=2, var=1, len_scale=10)
+srf = gs.SRF(model, seed=20170519)
+srf.structured([x, y])
+gs.transform.normal_to_lognormal(srf)
+srf.plot()
diff --git a/examples/07_transformations/01_binary.py b/examples/07_transformations/01_binary.py
new file mode 100755
index 00000000..20b41919
--- /dev/null
+++ b/examples/07_transformations/01_binary.py
@@ -0,0 +1,17 @@
+"""
+binary fields
+-------------
+
+Here we transform a field to a binary field with only two values.
+The dividing value is the mean by default and the upper and lower values
+are derived to preserve the variance.
+"""
+import gstools as gs
+
+# structured field with a size of 100x100 and a grid-size of 1x1
+x = y = range(100)
+model = gs.Gaussian(dim=2, var=1, len_scale=10)
+srf = gs.SRF(model, seed=20170519)
+srf.structured([x, y])
+gs.transform.binary(srf)
+srf.plot()
diff --git a/examples/07_transformations/02_discrete.py b/examples/07_transformations/02_discrete.py
new file mode 100755
index 00000000..43933537
--- /dev/null
+++ b/examples/07_transformations/02_discrete.py
@@ -0,0 +1,37 @@
+"""
+discrete fields
+-------------
+
+Here we transform a field to a discrete field with values.
+If we do not give thresholds, the pairwise means of the given
+values are taken as thresholds.
+If thresholds are given, arbitrary values can be applied to the field.
+"""
+import numpy as np
+import gstools as gs
+
+# structured field with a size of 100x100 and a grid-size of 0.5x0.5
+x = y = np.arange(200) * 0.5
+model = gs.Gaussian(dim=2, var=1, len_scale=5)
+srf = gs.SRF(model, seed=20170519)
+
+# create 5 equidistanly spaced values, thresholds are the arithmetic means
+srf.structured([x, y])
+discrete_values = np.linspace(np.min(srf.field), np.max(srf.field), 5)
+gs.transform.discrete(srf, discrete_values)
+srf.plot()
+
+# calculate thresholds for equal shares
+# but apply different values to the separated classes
+discrete_values2 = [0, -1, 2, -3, 4]
+srf.structured([x, y])
+gs.transform.discrete(srf, discrete_values2, thresholds="equal")
+srf.plot()
+
+# user defined thresholds
+thresholds = [-1, 1]
+# apply different values to the separated classes
+discrete_values3 = [0, 1, 10]
+srf.structured([x, y])
+gs.transform.discrete(srf, discrete_values3, thresholds=thresholds)
+srf.plot()
diff --git a/examples/07_transformations/03_zinn_harvey.py b/examples/07_transformations/03_zinn_harvey.py
new file mode 100755
index 00000000..d3d28a1a
--- /dev/null
+++ b/examples/07_transformations/03_zinn_harvey.py
@@ -0,0 +1,18 @@
+"""
+Zinn & Harvey transformation
+----------------------------
+
+Here, we transform a field with the so called "Zinn & Harvey" transformation presented in
+`Zinn & Harvey (2003) `__.
+With this transformation, one could overcome the restriction that in ordinary
+Gaussian random fields the mean values are the ones being the most connected.
+"""
+import gstools as gs
+
+# structured field with a size of 100x100 and a grid-size of 1x1
+x = y = range(100)
+model = gs.Gaussian(dim=2, var=1, len_scale=10)
+srf = gs.SRF(model, seed=20170519)
+srf.structured([x, y])
+gs.transform.zinnharvey(srf, conn="high")
+srf.plot()
diff --git a/examples/07_transformations/04_bimodal.py b/examples/07_transformations/04_bimodal.py
new file mode 100755
index 00000000..e0d4b2de
--- /dev/null
+++ b/examples/07_transformations/04_bimodal.py
@@ -0,0 +1,20 @@
+"""
+bimodal fields
+---------------
+
+We provide two transformations to obtain bimodal distributions:
+
+* `arcsin `__.
+* `uquad `__.
+
+Both transformations will preserve the mean and variance of the given field by default.
+"""
+import gstools as gs
+
+# structured field with a size of 100x100 and a grid-size of 1x1
+x = y = range(100)
+model = gs.Gaussian(dim=2, var=1, len_scale=10)
+srf = gs.SRF(model, seed=20170519)
+field = srf.structured([x, y])
+gs.transform.normal_to_arcsin(srf)
+srf.plot()
diff --git a/examples/07_transformations/05_combinations.py b/examples/07_transformations/05_combinations.py
new file mode 100755
index 00000000..2edb1a18
--- /dev/null
+++ b/examples/07_transformations/05_combinations.py
@@ -0,0 +1,29 @@
+"""
+Combinations
+------------
+
+You can combine different transformations simply by successively applying them.
+
+Here, we first force the single field realization to hold the given moments,
+namely mean and variance.
+Then we apply the Zinn & Harvey transformation to connect the low values.
+Afterwards the field is transformed to a binary field and last but not least,
+we transform it to log-values.
+"""
+import gstools as gs
+
+# structured field with a size of 100x100 and a grid-size of 1x1
+x = y = range(100)
+model = gs.Gaussian(dim=2, var=1, len_scale=10)
+srf = gs.SRF(model, mean=-9, seed=20170519)
+srf.structured([x, y])
+gs.transform.normal_force_moments(srf)
+gs.transform.zinnharvey(srf, conn="low")
+gs.transform.binary(srf)
+gs.transform.normal_to_lognormal(srf)
+srf.plot()
+
+###############################################################################
+# The resulting field could be interpreted as a transmissivity field, where
+# the values of low permeability are the ones being the most connected
+# and only two kinds of soil exist.
diff --git a/examples/07_transformations/README.rst b/examples/07_transformations/README.rst
new file mode 100644
index 00000000..d3a2ba06
--- /dev/null
+++ b/examples/07_transformations/README.rst
@@ -0,0 +1,37 @@
+Tutorial 7: Field transformations
+=================================
+
+The generated fields of gstools are ordinary Gaussian random fields.
+In application there are several transformations to describe real world
+problems in an appropriate manner.
+
+GStools provides a submodule :py:mod:`gstools.transform` with a range of
+common transformations:
+
+.. currentmodule:: gstools.transform
+
+.. autosummary::
+ binary
+ discrete
+ boxcox
+ zinnharvey
+ normal_force_moments
+ normal_to_lognormal
+ normal_to_uniform
+ normal_to_arcsin
+ normal_to_uquad
+
+
+All the transformations take a field class, that holds a generated field,
+as input and will manipulate this field inplace.
+
+Simply import the transform submodule and apply a transformation to the srf class:
+
+.. code-block:: python
+
+ from gstools import transform as tf
+ ...
+ tf.normal_to_lognormal(srf)
+
+Gallery
+-------
diff --git a/examples/08_variogram_estimation.py b/examples/08_variogram_estimation.py
deleted file mode 100644
index 167638d9..00000000
--- a/examples/08_variogram_estimation.py
+++ /dev/null
@@ -1,206 +0,0 @@
-import os
-from shutil import rmtree
-import zipfile
-import urllib.request
-import numpy as np
-import matplotlib.pyplot as pt
-from gstools import (
- vario_estimate_unstructured,
- vario_estimate_structured,
- Exponential,
-)
-from gstools.covmodel.plot import plot_variogram
-
-
-def download_herten():
- # download the data, warning: its about 250MB
- print("Downloading Herten data")
- data_filename = "data.zip"
- data_url = "http://store.pangaea.de/Publications/Bayer_et_al_2015/Herten-analog.zip"
- urllib.request.urlretrieve(data_url, "data.zip")
-
- with zipfile.ZipFile(data_filename, "r") as zf:
- zf.extract(
- os.path.join("Herten-analog", "sim-big_1000x1000x140", "sim.vtk")
- )
-
-
-def download_scripts():
- import fileinput
-
- # download a script for file conversion
- print("Downloading scripts")
- tools_filename = "scripts.zip"
- tool_url = (
- "http://store.pangaea.de/Publications/Bayer_et_al_2015/tools.zip"
- )
- urllib.request.urlretrieve(tool_url, tools_filename)
-
- filename = os.path.join("tools", "vtk2gslib.py")
-
- with zipfile.ZipFile(tools_filename, "r") as zf:
- zf.extract(filename)
-
- with fileinput.FileInput(filename, inplace=True) as fin:
- for line in fin:
- print(line.replace("header=-1", "header=None"), end="")
-
-
-def create_unstructured_grid(x_s, y_s):
- x_u, y_u = np.meshgrid(x_s, y_s)
- len_unstruct = len(x_s) * len(y_s)
- x_u = np.reshape(x_u, len_unstruct)
- y_u = np.reshape(y_u, len_unstruct)
- return x_u, y_u
-
-
-###############################################################################
-# data preparation ############################################################
-###############################################################################
-
-# uncomment these two function calls, in case the data was already downloaded
-# and you want to execute this script multiple times. But don't forget to
-# comment out the cleanup code at the end of this script.
-download_herten()
-download_scripts()
-
-# import the downloaded conversion script
-from tools.vtk2gslib import vtk2numpy
-
-# load the Herten aquifer with the downloaded vtk2numpy routine
-print("Loading data")
-herten, grid = vtk2numpy(
- os.path.join("Herten-analog", "sim-big_1000x1000x140", "sim.vtk")
-)
-
-# conductivity values per fazies from the supplementary data
-cond = np.array(
- [
- 2.50e-04,
- 2.30e-04,
- 6.10e-05,
- 2.60e-02,
- 1.30e-01,
- 9.50e-02,
- 4.30e-05,
- 6.00e-07,
- 2.30e-03,
- 1.40e-04,
- ]
-)
-
-# asign the conductivities to the facies
-herten_cond = cond[herten]
-
-# integrate over the vertical axis, calculate transmissivity
-herten_log_trans = np.log(np.sum(herten_cond, axis=2) * grid["dz"])
-
-# create a structured grid on which the data is defined
-x_s = np.arange(grid["ox"], grid["nx"] * grid["dx"], grid["dx"])
-y_s = np.arange(grid["oy"], grid["ny"] * grid["dy"], grid["dy"])
-
-pt.imshow(herten_log_trans.T, origin="lower", aspect="equal")
-pt.show()
-
-# create an unstructured grid for the variogram estimation
-x_u, y_u = create_unstructured_grid(x_s, y_s)
-
-###############################################################################
-# estimate the variogram on an unstructured grid ##############################
-###############################################################################
-
-bins = np.linspace(0, 10, 50)
-print("Estimating unstructured variogram")
-bin_center, gamma = vario_estimate_unstructured(
- (x_u, y_u),
- herten_log_trans.reshape(-1),
- bins,
- sampling_size=2000,
- sampling_seed=19920516,
-)
-
-# fit an exponential model
-fit_model = Exponential(dim=2)
-fit_model.fit_variogram(bin_center, gamma, nugget=False)
-
-pt.plot(bin_center, gamma)
-plot_variogram(fit_model, x_max=bins[-1])
-
-###############################################################################
-# estimate the variogram on a structured grid #################################
-###############################################################################
-
-# estimate the variogram on a structured grid
-# use only every 10th value, otherwise calculations would take very long
-x_s_skip = x_s[::10]
-y_s_skip = y_s[::10]
-herten_trans_skip = herten_log_trans[::10, ::10]
-
-print("Estimating structured variograms")
-gamma_x = vario_estimate_structured(herten_trans_skip, direction="x")
-gamma_y = vario_estimate_structured(herten_trans_skip, direction="y")
-
-x_plot = x_s_skip[:21]
-y_plot = y_s_skip[:21]
-# fit an exponential model
-fit_model_x = Exponential(dim=2)
-fit_model_x.fit_variogram(x_plot, gamma_x[:21], nugget=False)
-fit_model_y = Exponential(dim=2)
-fit_model_y.fit_variogram(y_plot, gamma_y[:21], nugget=False)
-
-line, = pt.plot(bin_center, gamma, label="estimated variogram (isotropic)")
-pt.plot(
- bin_center,
- fit_model.variogram(bin_center),
- color=line.get_color(),
- linestyle="--",
- label="exp. variogram (isotropic)",
-)
-
-line, = pt.plot(x_plot, gamma_x[:21], label="estimated variogram in x-dir")
-pt.plot(
- x_plot,
- fit_model_x.variogram(x_plot),
- color=line.get_color(),
- linestyle="--",
- label="exp. variogram in x-dir",
-)
-
-line, = pt.plot(y_plot, gamma_y[:21], label="estimated variogram in y-dir")
-pt.plot(
- y_plot,
- fit_model_y.variogram(y_plot),
- color=line.get_color(),
- linestyle="--",
- label="exp. variogram in y-dir",
-)
-
-pt.legend()
-pt.show()
-
-print("semivariogram model (isotropic):\n", fit_model)
-print("semivariogram model (in x-dir.):\n", fit_model_x)
-print("semivariogram model (in y-dir.):\n", fit_model_y)
-
-###############################################################################
-# creating a SRF from the Herten parameters ###################################
-###############################################################################
-
-from gstools import SRF
-
-srf = SRF(fit_model, seed=19770928)
-print("Calculating SRF")
-new_herten = srf((x_s, y_s), mesh_type="structured")
-
-pt.imshow(new_herten.T, origin="lower")
-pt.show()
-
-###############################################################################
-# cleanup #####################################################################
-###############################################################################
-
-# comment all in case you want to keep the data for playing around with it
-os.remove("data.zip")
-os.remove("scripts.zip")
-rmtree("Herten-analog")
-rmtree("tools")
diff --git a/examples/09_vector_field.py b/examples/09_vector_field.py
deleted file mode 100644
index 27c9cf6b..00000000
--- a/examples/09_vector_field.py
+++ /dev/null
@@ -1,21 +0,0 @@
-import numpy as np
-import matplotlib.pyplot as plt
-from gstools import SRF, Gaussian, Exponential
-
-# the grid
-x = np.arange(100)
-y = np.arange(100)
-
-# a smooth Gaussian covariance model
-model = Gaussian(dim=2, var=1, len_scale=10)
-
-srf = SRF(model, generator="VectorField")
-srf((x, y), mesh_type="structured", seed=19841203)
-srf.plot()
-
-# a rougher exponential covariance model
-model2 = Exponential(dim=2, var=1, len_scale=10)
-
-srf.model = model2
-srf((x, y), mesh_type="structured", seed=19841203)
-srf.plot()
diff --git a/examples/10_simple_kriging.py b/examples/10_simple_kriging.py
deleted file mode 100755
index 48c676d6..00000000
--- a/examples/10_simple_kriging.py
+++ /dev/null
@@ -1,15 +0,0 @@
-import numpy as np
-from gstools import Gaussian, krige
-
-# condtions
-cond_pos = [0.3, 1.9, 1.1, 3.3, 4.7]
-cond_val = [0.47, 0.56, 0.74, 1.47, 1.74]
-# resulting grid
-gridx = np.linspace(0.0, 15.0, 151)
-# spatial random field class
-model = Gaussian(dim=1, var=0.5, len_scale=2)
-krig = krige.Simple(model, mean=1, cond_pos=cond_pos, cond_val=cond_val)
-krig(gridx)
-ax = krig.plot()
-ax.scatter(cond_pos, cond_val, color="k", zorder=10, label="Conditions")
-ax.legend()
diff --git a/examples/11_ordinary_kriging.py b/examples/11_ordinary_kriging.py
deleted file mode 100644
index 2ea0af24..00000000
--- a/examples/11_ordinary_kriging.py
+++ /dev/null
@@ -1,15 +0,0 @@
-import numpy as np
-from gstools import Gaussian, krige
-
-# condtions
-cond_pos = [0.3, 1.9, 1.1, 3.3, 4.7]
-cond_val = [0.47, 0.56, 0.74, 1.47, 1.74]
-# resulting grid
-gridx = np.linspace(0.0, 15.0, 151)
-# spatial random field class
-model = Gaussian(dim=1, var=0.5, len_scale=2)
-krig = krige.Ordinary(model, cond_pos=cond_pos, cond_val=cond_val)
-krig(gridx)
-ax = krig.plot()
-ax.scatter(cond_pos, cond_val, color="k", zorder=10, label="Conditions")
-ax.legend()
diff --git a/examples/13_condition_ensemble.py b/examples/13_condition_ensemble.py
deleted file mode 100644
index 8730adcc..00000000
--- a/examples/13_condition_ensemble.py
+++ /dev/null
@@ -1,25 +0,0 @@
-import numpy as np
-from gstools import Gaussian, SRF
-import matplotlib.pyplot as plt
-
-# condtions
-cond_pos = [0.3, 1.9, 1.1, 3.3, 4.7]
-cond_val = [0.47, 0.56, 0.74, 1.47, 1.74]
-gridx = np.linspace(0.0, 15.0, 151)
-# spatial random field class
-model = Gaussian(dim=1, var=0.5, len_scale=2)
-srf = SRF(model)
-srf.set_condition(cond_pos, cond_val, "ordinary")
-fields = []
-for i in range(100):
- if i % 10 == 0:
- print(i)
- fields.append(srf(gridx, seed=i))
- label = "Conditioned ensemble" if i == 0 else None
- plt.plot(gridx, fields[i], color="k", alpha=0.1, label=label)
-plt.plot(gridx, np.full_like(gridx, srf.mean), label="estimated mean")
-plt.plot(gridx, np.mean(fields, axis=0), linestyle=":", label="Ensemble mean")
-plt.plot(gridx, srf.krige_field, linestyle="dashed", label="kriged field")
-plt.scatter(cond_pos, cond_val, color="k", zorder=10, label="Conditions")
-plt.legend()
-plt.show()
diff --git a/examples/14_transform_01.py b/examples/14_transform_01.py
deleted file mode 100755
index 28dc847f..00000000
--- a/examples/14_transform_01.py
+++ /dev/null
@@ -1,10 +0,0 @@
-from gstools import SRF, Gaussian
-from gstools import transform as tf
-
-# structured field with a size of 100x100 and a grid-size of 1x1
-x = y = range(100)
-model = Gaussian(dim=2, var=1, len_scale=10)
-srf = SRF(model, seed=20170519)
-srf.structured([x, y])
-tf.normal_to_lognormal(srf)
-srf.plot()
diff --git a/examples/15_transform_02.py b/examples/15_transform_02.py
deleted file mode 100755
index 7a24cdd7..00000000
--- a/examples/15_transform_02.py
+++ /dev/null
@@ -1,10 +0,0 @@
-from gstools import SRF, Gaussian
-from gstools import transform as tf
-
-# structured field with a size of 100x100 and a grid-size of 1x1
-x = y = range(100)
-model = Gaussian(dim=2, var=1, len_scale=10)
-srf = SRF(model, seed=20170519)
-srf.structured([x, y])
-tf.binary(srf)
-srf.plot()
diff --git a/examples/16_transform_03.py b/examples/16_transform_03.py
deleted file mode 100755
index c83f6e45..00000000
--- a/examples/16_transform_03.py
+++ /dev/null
@@ -1,10 +0,0 @@
-from gstools import SRF, Gaussian
-from gstools import transform as tf
-
-# structured field with a size of 100x100 and a grid-size of 1x1
-x = y = range(100)
-model = Gaussian(dim=2, var=1, len_scale=10)
-srf = SRF(model, seed=20170519)
-srf.structured([x, y])
-tf.zinnharvey(srf, conn="high")
-srf.plot()
diff --git a/examples/17_transform_04.py b/examples/17_transform_04.py
deleted file mode 100755
index e674c2c3..00000000
--- a/examples/17_transform_04.py
+++ /dev/null
@@ -1,10 +0,0 @@
-from gstools import SRF, Gaussian
-from gstools import transform as tf
-
-# structured field with a size of 100x100 and a grid-size of 1x1
-x = y = range(100)
-model = Gaussian(dim=2, var=1, len_scale=10)
-srf = SRF(model, seed=20170519)
-field = srf.structured([x, y])
-tf.normal_to_arcsin(srf)
-srf.plot()
diff --git a/examples/18_transform_05.py b/examples/18_transform_05.py
deleted file mode 100755
index 9be5bdaf..00000000
--- a/examples/18_transform_05.py
+++ /dev/null
@@ -1,13 +0,0 @@
-from gstools import SRF, Gaussian
-from gstools import transform as tf
-
-# structured field with a size of 100x100 and a grid-size of 1x1
-x = y = range(100)
-model = Gaussian(dim=2, var=1, len_scale=10)
-srf = SRF(model, mean=-9, seed=20170519)
-srf.structured([x, y])
-tf.normal_force_moments(srf)
-tf.zinnharvey(srf, conn="low")
-tf.binary(srf)
-tf.normal_to_lognormal(srf)
-srf.plot()
diff --git a/examples/20_pykrige_interface.py b/examples/20_pykrige_interface.py
deleted file mode 100755
index 716cacb8..00000000
--- a/examples/20_pykrige_interface.py
+++ /dev/null
@@ -1,33 +0,0 @@
-# -*- coding: utf-8 -*-
-"""Example how to use the PyKrige routines with a GSTools CovModel."""
-import numpy as np
-from gstools import Gaussian, krige
-from pykrige.ok import OrdinaryKriging
-from matplotlib import pyplot as plt
-
-# conditioning data
-data = np.array(
- [
- [0.3, 1.2, 0.47],
- [1.9, 0.6, 0.56],
- [1.1, 3.2, 0.74],
- [3.3, 4.4, 1.47],
- [4.7, 3.8, 1.74],
- ]
-)
-# grid definition for output field
-gridx = np.arange(0.0, 5.5, 0.1)
-gridy = np.arange(0.0, 6.5, 0.1)
-# a GSTools based covariance model
-cov_model = Gaussian(
- dim=2, len_scale=1, anis=0.2, angles=-0.5, var=0.5, nugget=0.1
-)
-# ordinary kriging with pykrige
-OK1 = OrdinaryKriging(data[:, 0], data[:, 1], data[:, 2], cov_model)
-z1, ss1 = OK1.execute("grid", gridx, gridy)
-plt.imshow(z1, origin="lower")
-plt.show()
-# ordinary kriging with gstools for comparison
-OK2 = krige.Ordinary(cov_model, [data[:, 0], data[:, 1]], data[:, 2])
-OK2.structured([gridx, gridy])
-OK2.plot()
diff --git a/gstools/__init__.py b/gstools/__init__.py
index c10c8da3..cc6a7d61 100644
--- a/gstools/__init__.py
+++ b/gstools/__init__.py
@@ -81,12 +81,12 @@
.. currentmodule:: gstools.tools
.. autosummary::
- to_vtk
vtk_export
- to_vtk_structured
vtk_export_structured
- to_vtk_unstructured
vtk_export_unstructured
+ to_vtk
+ to_vtk_structured
+ to_vtk_unstructured
variogram estimation
^^^^^^^^^^^^^^^^^^^^
@@ -99,17 +99,15 @@
vario_estimate_unstructured
"""
-from __future__ import absolute_import
-
-import sys
-
-from gstools._version import __version__
from gstools import field, variogram, random, covmodel, tools, krige, transform
from gstools.field import SRF
-from gstools.tools.export import (
+from gstools.tools import (
+ vtk_export,
vtk_export_structured,
vtk_export_unstructured,
- vtk_export,
+ to_vtk,
+ to_vtk_structured,
+ to_vtk_unstructured,
)
from gstools.variogram import (
vario_estimate_structured,
@@ -131,20 +129,11 @@
TPLStable,
)
-
-PY_VERSION = sys.version_info
-DEPRECATION_STR = (
- "DEPRECATION: Python {0} will reach the end of is life on "
- "{1}. Please upgrade your Python as Python {0} "
- "won't be maintained after that date. A future version of GSTools will "
- "drop support for Python {0}."
-)
-
-if PY_VERSION[:2] == (2, 7):
- print(DEPRECATION_STR.format(2.7, "1st January 2020"))
-elif PY_VERSION[:2] == (3, 4):
- print(DEPRECATION_STR.format(3.4, "18th March 2019"))
-
+try:
+ from gstools._version import __version__
+except ModuleNotFoundError: # pragma: nocover
+ # package is not installed
+ __version__ = "0.0.0.dev0"
__all__ = ["__version__"]
__all__ += ["covmodel", "field", "variogram", "krige", "random", "tools"]
@@ -169,10 +158,10 @@
__all__ += [
"SRF",
- "to_vtk_structured",
+ "vtk_export",
"vtk_export_structured",
- "to_vtk_unstructured",
"vtk_export_unstructured",
"to_vtk",
- "vtk_export",
+ "to_vtk_structured",
+ "to_vtk_unstructured",
]
diff --git a/gstools/_version.py b/gstools/_version.py
deleted file mode 100644
index ac10cb3d..00000000
--- a/gstools/_version.py
+++ /dev/null
@@ -1,2 +0,0 @@
-"""Provide a central version."""
-__version__ = "1.1.1"
diff --git a/gstools/covmodel/__init__.py b/gstools/covmodel/__init__.py
index cf30d3ff..f638f2cf 100644
--- a/gstools/covmodel/__init__.py
+++ b/gstools/covmodel/__init__.py
@@ -2,33 +2,30 @@
"""
GStools subpackage providing a set of handy covariance models.
+.. currentmodule:: gstools.covmodel
+
Subpackages
^^^^^^^^^^^
-.. currentmodule:: gstools.covmodel
-
.. autosummary::
- base
- models
- tpl_models
plot
Covariance Base-Class
^^^^^^^^^^^^^^^^^^^^^
Class to construct user defined covariance models
-.. currentmodule:: gstools.covmodel.base
-
.. autosummary::
+ :toctree: generated
+
CovModel
Covariance Models
^^^^^^^^^^^^^^^^^
Standard Covariance Models
-.. currentmodule:: gstools.covmodel.models
-
.. autosummary::
+ :toctree: generated
+
Gaussian
Exponential
Matern
@@ -41,14 +38,13 @@
Truncated Power Law Covariance Models
-.. currentmodule:: gstools.covmodel.tpl_models
-
.. autosummary::
+ :toctree: generated
+
TPLGaussian
TPLExponential
TPLStable
"""
-from __future__ import absolute_import
from gstools.covmodel.base import CovModel
from gstools.covmodel.models import (
diff --git a/gstools/covmodel/base.py b/gstools/covmodel/base.py
index 45d2362e..cbac3892 100644
--- a/gstools/covmodel/base.py
+++ b/gstools/covmodel/base.py
@@ -10,9 +10,9 @@
CovModel
"""
# pylint: disable=C0103, R0201
-from __future__ import print_function, division, absolute_import
-import six
+import warnings
+import copy
import numpy as np
from scipy.integrate import quad as integral
from scipy.optimize import curve_fit, root
@@ -31,17 +31,17 @@
__all__ = ["CovModel"]
# default arguments for hankel.SymmetricFourierTransform
-HANKEL_DEFAULT = {
- "a": -1, # should only be changed, if you know exactly what
- "b": 1, # you do or if you are crazy
- "N": 1000,
- "h": 0.001,
-}
+HANKEL_DEFAULT = {"a": -1, "b": 1, "N": 200, "h": 0.001, "alt": True}
+
+
+class AttributeWarning(UserWarning):
+ pass
+
# The CovModel Base-Class #####################################################
-class CovModel(six.with_metaclass(InitSubclassMeta)):
+class CovModel(metaclass=InitSubclassMeta):
r"""Base class for the GSTools covariance models.
Parameters
@@ -56,15 +56,22 @@ class CovModel(six.with_metaclass(InitSubclassMeta)):
If a single value is given, the same length-scale will be used for
every direction. If multiple values (for main and transversal
directions) are given, `anis` will be
- recalculated accordingly.
+ recalculated accordingly. If only two values are given in 3D,
+ the latter one will be used for both transversal directions.
Default: ``1.0``
nugget : :class:`float`, optional
nugget of the model. Default: ``0.0``
anis : :class:`float` or :class:`list`, optional
- anisotropy ratios in the transversal directions [y, z].
+ anisotropy ratios in the transversal directions [e_y, e_z].
+
+ * e_y = l_y / l_x
+ * e_z = l_z / l_x
+
+ If only one value is given in 3D, e_y will be set to 1.
+ This value will be ignored, if multiple len_scales are given.
Default: ``1.0``
angles : :class:`float` or :class:`list`, optional
- angles of rotation:
+ angles of rotation (given in rad):
* in 2D: given as rotation around z-axis
* in 3D: given by yaw, pitch, and roll (known as Tait–Bryan angles)
@@ -137,6 +144,13 @@ def __init__(
+ "' has a 'bad' name, since it is already present in "
+ "the class. It could not be added to the model"
)
+ if opt_name not in self.default_opt_arg().keys():
+ warnings.warn(
+ "The given optional argument '{}' ".format(opt_name)
+ + "is unknown or has at least no defined standard value. "
+ + "Or you made a Typo... hehe.",
+ AttributeWarning,
+ )
# Magic happens here
setattr(self, opt_name, opt_arg[opt_name])
@@ -196,25 +210,25 @@ def __init_subclass__(cls):
* ``model.variogram(r)``
:math:`\gamma\left(r\right)=
- \sigma^2\cdot\left(1-\mathrm{cor}\left(r\right)\right)+n`
+ \sigma^2\cdot\left(1-\rho\left(r\right)\right)+n`
* ``model.covariance(r)``
:math:`C\left(r\right)=
- \sigma^2\cdot\mathrm{cor}\left(r\right)`
+ \sigma^2\cdot\rho\left(r\right)`
* ``model.correlation(r)``
- :math:`\mathrm{cor}\left(r\right)`
+ :math:`\rho\left(r\right)`
Best practice is to use the ``correlation`` function, or the ``cor``
function. The latter one takes the dimensionles distance h=r/l.
"""
- # overrid one of these ################################################
+ # override one of these ###############################################
def variogram(self, r):
r"""Isotropic variogram of the model.
Given by: :math:`\gamma\left(r\right)=
- \sigma^2\cdot\left(1-\mathrm{cor}\left(r\right)\right)+n`
+ \sigma^2\cdot\left(1-\rho\left(r\right)\right)+n`
- Where :math:`\mathrm{cor}(r)` is the correlation function.
+ Where :math:`\rho(r)` is the correlation function.
"""
return self.var - self.covariance(r) + self.nugget
@@ -222,39 +236,49 @@ def covariance(self, r):
r"""Covariance of the model.
Given by: :math:`C\left(r\right)=
- \sigma^2\cdot\mathrm{cor}\left(r\right)`
+ \sigma^2\cdot\rho\left(r\right)`
- Where :math:`\mathrm{cor}(r)` is the correlation function.
+ Where :math:`\rho(r)` is the correlation function.
"""
return self.var * self.correlation(r)
def correlation(self, r):
r"""Correlation function (or normalized covariance) of the model.
- Given by: :math:`\mathrm{cor}\left(r\right)`
+ Given by: :math:`\rho\left(r\right)`
It has to be a monotonic decreasing function with
- :math:`\mathrm{cor}(0)=1` and :math:`\mathrm{cor}(\infty)=0`.
+ :math:`\rho(0)=1` and :math:`\rho(\infty)=0`.
"""
return 1.0 - (self.variogram(r) - self.nugget) / self.var
- def cor_from_cor(self, r):
+ def correlation_from_cor(self, r):
r"""Correlation function (or normalized covariance) of the model.
- Given by: :math:`\mathrm{cor}\left(r\right)`
+ Given by: :math:`\rho\left(r\right)`
It has to be a monotonic decreasing function with
- :math:`\mathrm{cor}(0)=1` and :math:`\mathrm{cor}(\infty)=0`.
+ :math:`\rho(0)=1` and :math:`\rho(\infty)=0`.
"""
r = np.array(np.abs(r), dtype=np.double)
return self.cor(r / self.len_scale)
+ def cor_from_correlation(self, h):
+ r"""Normalziled correlation function taking a normalized range.
+
+ Given by: :math:`\mathrm{cor}\left(r/\ell\right) = \rho(r)`
+ """
+ h = np.array(np.abs(h), dtype=np.double)
+ return self.correlation(h * self.len_scale)
+
#######################################################################
abstract = True
if hasattr(cls, "cor"):
- cls.correlation = cor_from_cor
+ cls.correlation = correlation_from_cor
abstract = False
+ else:
+ cls.cor = cor_from_correlation
if not hasattr(cls, "variogram"):
cls.variogram = variogram
else:
@@ -304,7 +328,8 @@ def _get_iso_rad(self, pos):
x, y, z = pos2xyz(pos, max_dim=self.dim)
if self.do_rotation:
x, y, z = unrotate_mesh(self.dim, self.angles, x, y, z)
- y, z = make_isotropic(self.dim, self.anis, y, z)
+ if not self.is_isotropic:
+ y, z = make_isotropic(self.dim, self.anis, y, z)
return np.linalg.norm((x, y, z)[: self.dim], axis=0)
def vario_spatial(self, pos):
@@ -323,9 +348,9 @@ def cov_nugget(self, r):
r"""Covariance of the model respecting the nugget at r=0.
Given by: :math:`C\left(r\right)=
- \sigma^2\cdot\mathrm{cor}\left(r\right)`
+ \sigma^2\cdot\rho\left(r\right)`
- Where :math:`\mathrm{cor}(r)` is the correlation function.
+ Where :math:`\rho(r)` is the correlation function.
"""
r = np.array(np.abs(r), dtype=np.double)
r_gz = np.logical_not(np.isclose(r, 0))
@@ -338,9 +363,9 @@ def vario_nugget(self, r):
r"""Isotropic variogram of the model respecting the nugget at r=0.
Given by: :math:`\gamma\left(r\right)=
- \sigma^2\cdot\left(1-\mathrm{cor}\left(r\right)\right)+n`
+ \sigma^2\cdot\left(1-\rho\left(r\right)\right)+n`
- Where :math:`\mathrm{cor}(r)` is the correlation function.
+ Where :math:`\rho(r)` is the correlation function.
"""
r = np.array(np.abs(r), dtype=np.double)
r_gz = np.logical_not(np.isclose(r, 0))
@@ -387,9 +412,9 @@ def pykrige_vario(self, args=None, r=0):
r"""Isotropic variogram of the model for pykrige.
Given by: :math:`\gamma\left(r\right)=
- \sigma^2\cdot\left(1-\mathrm{cor}\left(r\right)\right)+n`
+ \sigma^2\cdot\left(1-\rho\left(r\right)\right)+n`
- Where :math:`\mathrm{cor}(r)` is the correlation function.
+ Where :math:`\rho(r)` is the correlation function.
"""
return self.variogram(r)
@@ -577,24 +602,7 @@ def spectral_density(self, k):
Radius of the phase: :math:`k=\left\Vert\mathbf{k}\right\Vert`
"""
k = np.array(np.abs(k), dtype=np.double)
- if self.dim > 1:
- res = self._sft.transform(self.correlation, k, ret_err=False)
- else:
- k_gz = np.logical_not(np.isclose(k, 0))
- res = np.empty_like(k, dtype=np.double)
- res[k_gz] = self._sft.transform(
- self.correlation, k[k_gz], ret_err=False
- )
- # this is a hack for k=0, we calculate by hand
- fac = (
- np.sqrt(
- np.abs(self.hankel_kw["b"])
- / (2 * np.pi) ** (1 - self.hankel_kw["a"])
- )
- * 2
- )
- res[np.logical_not(k_gz)] = self.integral_scale * fac
- return res
+ return self._sft.transform(self.correlation, k, ret_err=False)
def spectral_rad_pdf(self, r):
"""Radial spectral density of the model."""
@@ -616,11 +624,8 @@ def spectral_rad_pdf(self, r):
def ln_spectral_rad_pdf(self, r):
"""Log radial spectral density of the model."""
- spec = np.array(self.spectral_rad_pdf(r))
- spec_gz = np.logical_not(np.isclose(spec, 0))
- res = np.full_like(spec, -np.inf, dtype=np.double)
- res[spec_gz] = np.log(spec[spec_gz])
- return res
+ with np.errstate(divide="ignore"):
+ return np.log(self.spectral_rad_pdf(r))
def _has_cdf(self):
"""State if a cdf is defined with 'spectral_rad_cdf'."""
@@ -844,7 +849,7 @@ def check_arg_bounds(self):
+ str(val)
)
- ### bounds properties ####################################################
+ ### bounds properties #####################################################
@property
def var_bounds(self):
@@ -1092,12 +1097,13 @@ def hankel_kw(self):
@hankel_kw.setter
def hankel_kw(self, hankel_kw):
- self._hankel_kw = HANKEL_DEFAULT if hankel_kw is None else hankel_kw
+ if self._hankel_kw is None or hankel_kw is None:
+ self._hankel_kw = copy.copy(HANKEL_DEFAULT)
+ if hankel_kw is not None:
+ self._hankel_kw.update(hankel_kw)
if self.dim is not None:
self._sft = SFT(ndim=self.dim, **self.hankel_kw)
- ### properties ############################################################
-
@property
def dist_func(self):
""":class:`tuple` of :any:`callable`: pdf, cdf and ppf.
@@ -1186,9 +1192,14 @@ def name(self):
@property
def do_rotation(self):
""":any:`bool`: State if a rotation is performed."""
- return not np.all(np.isclose(self.angles, 0.0))
+ return (
+ not np.all(np.isclose(self.angles, 0.0)) and not self.is_isotropic
+ )
- ### magic methods #########################################################
+ @property
+ def is_isotropic(self):
+ """:any:`bool`: State if a model is isotropic."""
+ return np.all(np.isclose(self.anis, 1.0))
def __eq__(self, other):
"""Compare CovModels."""
diff --git a/gstools/covmodel/models.py b/gstools/covmodel/models.py
index 6a748ba6..171f1bfc 100644
--- a/gstools/covmodel/models.py
+++ b/gstools/covmodel/models.py
@@ -18,12 +18,11 @@
Intersection
"""
# pylint: disable=C0103, E1101, E1137
-from __future__ import print_function, division, absolute_import
import warnings
import numpy as np
from scipy import special as sps
-from gstools.covmodel.base import CovModel
+from gstools.covmodel import CovModel
__all__ = [
"Gaussian",
@@ -49,7 +48,7 @@ class Gaussian(CovModel):
This model is given by the following correlation function:
.. math::
- \mathrm{cor}(r) =
+ \rho(r) =
\exp\left(- \frac{\pi}{4} \cdot \left(\frac{r}{\ell}\right)^2\right)
"""
@@ -58,7 +57,7 @@ def correlation(self, r):
r"""Gaussian correlation function.
.. math::
- \mathrm{cor}(r) =
+ \rho(r) =
\exp\left(- \frac{\pi}{4}\cdot \left(\frac{r}{\ell}\right)^2\right)
"""
r = np.array(np.abs(r), dtype=np.double)
@@ -101,9 +100,7 @@ def spectral_rad_ppf(self, u):
def _has_ppf(self):
# since the ppf is not analytical for dim=3, we have to state that
- if self.dim == 3:
- return False
- return True
+ return False if self.dim == 3 else True
def calc_integral_scale(self): # noqa: D102
return self.len_scale
@@ -120,7 +117,7 @@ class Exponential(CovModel):
This model is given by the following correlation function:
.. math::
- \mathrm{cor}(r) =
+ \rho(r) =
\exp\left(- \frac{r}{\ell} \right)
"""
@@ -129,7 +126,7 @@ def correlation(self, r):
r"""Exponential correlation function.
.. math::
- \mathrm{cor}(r) =
+ \rho(r) =
\exp\left(- \frac{r}{\ell} \right)
"""
r = np.array(np.abs(r), dtype=np.double)
@@ -184,9 +181,7 @@ def spectral_rad_ppf(self, u):
def _has_ppf(self):
# since the ppf is not analytical for dim=3, we have to state that
- if self.dim == 3:
- return False
- return True
+ return False if self.dim == 3 else True
def calc_integral_scale(self): # noqa: D102
return self.len_scale
@@ -203,7 +198,7 @@ class Rational(CovModel):
This model is given by the following correlation function:
.. math::
- \mathrm{cor}(r) =
+ \rho(r) =
\left(1 + \frac{1}{2\alpha} \cdot
\left(\frac{r}{\ell}\right)^2\right)^{-\alpha}
@@ -246,7 +241,7 @@ def correlation(self, r):
r"""Rational correlation function.
.. math::
- \mathrm{cor}(r) =
+ \rho(r) =
\left(1 + \frac{1}{2\alpha} \cdot
\left(\frac{r}{\ell}\right)^2\right)^{-\alpha}
"""
@@ -267,7 +262,7 @@ class Stable(CovModel):
This model is given by the following correlation function:
.. math::
- \mathrm{cor}(r) =
+ \rho(r) =
\exp\left(- \left(\frac{r}{\ell}\right)^{\alpha}\right)
:math:`\alpha` is a shape parameter with :math:`\alpha\in(0,2]`
@@ -324,7 +319,7 @@ def correlation(self, r):
r"""Stable correlation function.
.. math::
- \mathrm{cor}(r) =
+ \rho(r) =
\exp\left(- \left(\frac{r}{\ell}\right)^{\alpha}\right)
"""
r = np.array(np.abs(r), dtype=np.double)
@@ -342,7 +337,7 @@ class Matern(CovModel):
This model is given by the following correlation function:
.. math::
- \mathrm{cor}(r) =
+ \rho(r) =
\frac{2^{1-\nu}}{\Gamma\left(\nu\right)} \cdot
\left(\sqrt{\nu}\cdot\frac{r}{\ell}\right)^{\nu} \cdot
\mathrm{K}_{\nu}\left(\sqrt{\nu}\cdot\frac{r}{\ell}\right)
@@ -356,7 +351,7 @@ class Matern(CovModel):
case:
.. math::
- \mathrm{cor}(r) =
+ \rho(r) =
\exp\left(- \frac{1}{4} \cdot \left(\frac{r}{\ell}\right)^2\right)
Other Parameters
@@ -396,7 +391,7 @@ def correlation(self, r):
r"""Matérn correlation function.
.. math::
- \mathrm{cor}(r) =
+ \rho(r) =
\frac{2^{1-\nu}}{\Gamma\left(\nu\right)} \cdot
\left(\sqrt{\nu}\cdot\frac{r}{\ell}\right)^{\nu} \cdot
\mathrm{K}_{\nu}\left(\sqrt{\nu}\cdot\frac{r}{\ell}\right)
@@ -465,7 +460,7 @@ class Linear(CovModel):
This model is given by the following correlation function:
.. math::
- \mathrm{cor}(r) =
+ \rho(r) =
\begin{cases}
1-\frac{r}{\ell}
& r<\ell\\
@@ -478,7 +473,7 @@ def correlation(self, r):
r"""Linear correlation function.
.. math::
- \mathrm{cor}(r) =
+ \rho(r) =
\begin{cases}
1-\frac{r}{\ell}
& r<\ell\\
@@ -508,7 +503,7 @@ class Circular(CovModel):
This model is given by the following correlation function:
.. math::
- \mathrm{cor}(r) =
+ \rho(r) =
\begin{cases}
\frac{2}{\pi}\cdot\left(
\cos^{-1}\left(\frac{r}{\ell}\right) -
@@ -524,7 +519,7 @@ def correlation(self, r):
r"""Circular correlation function.
.. math::
- \mathrm{cor}(r) =
+ \rho(r) =
\begin{cases}
\frac{2}{\pi}\cdot\left(
\cos^{-1}\left(\frac{r}{\ell}\right) -
@@ -567,7 +562,7 @@ class Spherical(CovModel):
This model is given by the following correlation function:
.. math::
- \mathrm{cor}(r) =
+ \rho(r) =
\begin{cases}
1-\frac{3}{2}\cdot\frac{r}{\ell} +
\frac{1}{2}\cdot\left(\frac{r}{\ell}\right)^{3}
@@ -581,7 +576,7 @@ def correlation(self, r):
r"""Spherical correlation function.
.. math::
- \mathrm{cor}(r) =
+ \rho(r) =
\begin{cases}
1-\frac{3}{2}\cdot\frac{r}{\ell} +
\frac{1}{2}\cdot\left(\frac{r}{\ell}\right)^{3}
@@ -619,7 +614,7 @@ class Intersection(CovModel):
In 1D:
.. math::
- \mathrm{cor}(r) =
+ \rho(r) =
\begin{cases}
1-\frac{r}{\ell}
& r<\ell\\
@@ -629,7 +624,7 @@ class Intersection(CovModel):
In 2D:
.. math::
- \mathrm{cor}(r) =
+ \rho(r) =
\begin{cases}
\frac{2}{\pi}\cdot\left(
\cos^{-1}\left(\frac{r}{\ell}\right) -
@@ -642,7 +637,7 @@ class Intersection(CovModel):
In 3D:
.. math::
- \mathrm{cor}(r) =
+ \rho(r) =
\begin{cases}
1-\frac{3}{2}\cdot\frac{r}{\ell} +
\frac{1}{2}\cdot\left(\frac{r}{\ell}\right)^{3}
diff --git a/gstools/covmodel/plot.py b/gstools/covmodel/plot.py
index 1695c88a..c2a3f68b 100644
--- a/gstools/covmodel/plot.py
+++ b/gstools/covmodel/plot.py
@@ -18,7 +18,6 @@
plot_spectral_rad_pdf
"""
# pylint: disable=C0103
-from __future__ import print_function, division, absolute_import
import numpy as np
import gstools
@@ -62,6 +61,7 @@ def plot_vario_spatial(
): # pragma: no cover
"""Plot spatial variogram of a given CovModel."""
field = gstools.field.base.Field(model)
+ field._value_type = "scalar"
if x_max is None:
x_max = 3 * model.integral_scale
field.mesh_type = "structured"
@@ -79,6 +79,7 @@ def plot_cov_spatial(
): # pragma: no cover
"""Plot spatial covariance of a given CovModel."""
field = gstools.field.base.Field(model)
+ field._value_type = "scalar"
if x_max is None:
x_max = 3 * model.integral_scale
field.mesh_type = "structured"
@@ -96,6 +97,7 @@ def plot_cor_spatial(
): # pragma: no cover
"""Plot spatial correlation of a given CovModel."""
field = gstools.field.base.Field(model)
+ field._value_type = "scalar"
if x_max is None:
x_max = 3 * model.integral_scale
field.mesh_type = "structured"
diff --git a/gstools/covmodel/tools.py b/gstools/covmodel/tools.py
index 1a1e15c7..953631fc 100644
--- a/gstools/covmodel/tools.py
+++ b/gstools/covmodel/tools.py
@@ -15,9 +15,8 @@
exp_int
inc_beta
"""
-# pylint: disable=C0103
-from __future__ import print_function, division, absolute_import
+# pylint: disable=C0103
import numpy as np
from scipy import special as sps
@@ -111,7 +110,15 @@ def set_len_anis(dim, len_scale, anis):
Notes
-----
- If ``len_scale`` is given as list, ``anis`` will be recalculated.
+ If ``len_scale`` is given by at least two values,
+ ``anis`` will be recalculated.
+
+ If ``len_scale`` is given as list with to few values, the latter value will
+ be used for the remaining dimensions. (e.g. [l_1, l_2] in 3D is equal to
+ [l_1, l_2, l_2])
+
+ If to few ``anis`` values are given, the first dimensions will be filled
+ up with 1. (eg. anis=[e] in 3D is equal to anis=[1, e])
"""
ls_tmp = np.atleast_1d(len_scale)[:dim]
# use just one length scale (x-direction)
@@ -123,22 +130,14 @@ def set_len_anis(dim, len_scale, anis):
# fill up the anisotropies with ones, such that len()==dim-1
out_anis = np.pad(
out_anis,
- (0, dim - len(out_anis) - 1),
+ (dim - len(out_anis) - 1, 0),
"constant",
constant_values=1.0,
)
- elif dim == 1:
- # there is no anisotropy in 1 dimension
- out_anis = np.empty(0)
else:
# fill up length-scales with main len_scale, such that len()==dim
if len(ls_tmp) < dim:
- ls_tmp = np.pad(
- ls_tmp,
- (0, dim - len(ls_tmp)),
- "constant",
- constant_values=out_len_scale,
- )
+ ls_tmp = np.pad(ls_tmp, (0, dim - len(ls_tmp)), "edge")
# if multiple length-scales are given, calculate the anisotropies
out_anis = np.zeros(dim - 1, dtype=np.double)
for i in range(1, dim):
diff --git a/gstools/covmodel/tpl_models.py b/gstools/covmodel/tpl_models.py
index 4eae9aa9..27a9e0dc 100644
--- a/gstools/covmodel/tpl_models.py
+++ b/gstools/covmodel/tpl_models.py
@@ -12,12 +12,16 @@
TPLStable
"""
# pylint: disable=C0103, E1101
-from __future__ import print_function, division, absolute_import
import warnings
import numpy as np
from gstools.covmodel.base import CovModel
-from gstools.tools.special import tplstable_cor
+from gstools.tools.special import (
+ tplstable_cor,
+ tpl_gau_spec_dens,
+ tpl_exp_spec_dens,
+)
+
__all__ = ["TPLGaussian", "TPLExponential", "TPLStable"]
@@ -75,13 +79,13 @@ class TPLGaussian(CovModel):
The following Parameters occure:
- * :math:`C>0` : The scaling factor from the Power-Law.
+ * :math:`C>0` : scaling factor from the Power-Law
This parameter will be calculated internally by the given variance.
You can access C directly by ``model.var_raw``
- * :math:`00` : The scaling factor from the Power-Law.
+ * :math:`C>0` : scaling factor from the Power-Law
This parameter will be calculated internally by the given variance.
You can access C directly by ``model.var_raw``
- * :math:`00` : The scaling factor from the Power-Law.
+ * :math:`C>0` : scaling factor from the Power-Law
This parameter will be calculated internally by the given variance.
You can access C directly by ``model.var_raw``
- * :math:`0 2 or ext_drift.shape[1] != point_no:
+ raise ValueError("Krige: wrong number of cond. drifts.")
+ return ext_drift
+ ext_drift = np.array(ext_drift, dtype=np.double, ndmin=2)
+ ext_shape = np.shape(ext_drift)
+ shape = (self.drift_no, point_no)
+ if self.drift_no > 1 and ext_shape[0] != self.drift_no:
+ raise ValueError("Krige: wrong number of external drifts.")
+ if np.prod(ext_shape) != np.prod(shape):
+ raise ValueError("Krige: wrong number of ext. drift values.")
+ return np.array(ext_drift, dtype=np.double).reshape(shape)
+ elif not set_cond and self._cond_ext_drift.size > 0:
+ raise ValueError("Krige: wrong number of ext. drift values.")
+ return np.array([])
+
+ def _post_field(self, field, krige_var):
+ """
+ Postprocessing and saving of kriging field and error variance.
+
+ Parameters
+ ----------
+ field : :class:`numpy.ndarray`
+ Raw kriging field.
+ krige_var : :class:`numpy.ndarray`
+ Raw kriging error variance.
+ """
+ if self.trend_function is no_trend:
+ self.field = field
+ else:
+ self.field = field + eval_func(
+ self.trend_function, self.pos, self.mesh_type
+ )
+ self.krige_var = krige_var
+
+ def _get_dists(self, pos1, pos2=None, pos2_slice=(0, None)):
+ """
+ Calculate pairwise distances.
+
+ Parameters
+ ----------
+ pos1 : :class:`tuple` of :class:`numpy.ndarray`
+ the first position tuple
+ pos2 : :class:`tuple` of :class:`numpy.ndarray`, optional
+ the second position tuple. If none, the first one is taken.
+ pos2_slice : :class:`tuple` of :class:`int`, optional
+ Start and stop of slice for the pos2 array. Default: all values.
+
+ Returns
+ -------
+ :class:`numpy.ndarray`
+ Matrix containing the pairwise distances.
+ """
+ pos1_stack = np.column_stack(pos1[: self.model.dim])
+ if pos2 is None:
+ return cdist(pos1_stack, pos1_stack)
+ p2s = slice(*pos2_slice)
+ pos2_stack = np.column_stack(pos2[: self.model.dim])[p2s, ...]
+ return cdist(pos1_stack, pos2_stack)
+
+ def get_mean(self):
+ """Calculate the estimated mean."""
+ return self._mean
+
+ def set_condition(self, cond_pos, cond_val, ext_drift=None):
+ """Set the conditions for kriging.
+
+ Parameters
+ ----------
+ cond_pos : :class:`list`
+ the position tuple of the conditions (x, [y, z])
+ cond_val : :class:`numpy.ndarray`
+ the values of the conditions
+ ext_drift : :class:`numpy.ndarray` or :any:`None`, optional
+ the external drift values at the given conditions (only for EDK)
+ For multiple external drifts, the first dimension
+ should be the index of the drift term.
+ """
+ self._cond_pos, self._cond_val = set_condition(
+ cond_pos, cond_val, self.model.dim
+ )
+ self._cond_ext_drift = self._pre_ext_drift(
+ self.cond_no, ext_drift, set_cond=True
+ )
+ self.update()
+
+ def set_drift_functions(self, drift_functions=None):
+ """
+ Set the drift functions for universal kriging.
+
+ Parameters
+ ----------
+ drift_functions : :class:`list` of :any:`callable`, :class:`str` or :class:`int`
+ Either a list of callable functions, an integer representing
+ the polynomial order of the drift or one of the following strings:
+
+ * "linear" : regional linear drift (equals order=1)
+ * "quadratic" : regional quadratic drift (equals order=2)
+
+ Raises
+ ------
+ ValueError
+ If the given drift functions are not callable.
+ """
+ if drift_functions is None:
+ self._drift_functions = []
+ elif isinstance(drift_functions, (str, int)):
+ self._drift_functions = get_drift_functions(
+ self.model.dim, drift_functions
+ )
+ else:
+ if isinstance(drift_functions, collections.abc.Iterator):
+ drift_functions = list(drift_functions)
+ # check for a single content thats not a string
+ try:
+ iter(drift_functions)
+ except TypeError:
+ drift_functions = [drift_functions]
+ for f in drift_functions:
+ if not callable(f):
+ raise ValueError("Universal: Drift functions not callable")
+ self._drift_functions = drift_functions
+
+ def update(self):
+ """Update the kriging settings."""
+ x, y, z, __, __, __, __ = self._pre_pos(self.cond_pos)
+ # krige pos are the unrotated and isotropic condition positions
+ self._krige_pos = (x, y, z)[: self.model.dim]
+ self._krige_mat = self._get_krige_mat()
+ if self.trend_function is no_trend:
+ self._cond_trend = 0.0
+ else:
+ self._cond_trend = self.trend_function(*self.cond_pos)
+ self._mean = self.get_mean()
+
+ @property
+ def _krige_cond(self):
+ """:class:`numpy.ndarray`: The prepared kriging conditions."""
+ pad_size = self.drift_no + int(self.unbiased)
+ return np.pad(
+ self.cond_val - self.cond_trend,
+ (0, pad_size),
+ mode="constant",
+ constant_values=0,
+ )
+
+ @property
+ def cond_pos(self):
+ """:class:`list`: The position tuple of the conditions."""
+ return self._cond_pos
+
+ @property
+ def cond_val(self):
+ """:class:`list`: The values of the conditions."""
+ return self._cond_val
+
+ @property
+ def cond_no(self):
+ """:class:`int`: The number of the conditions."""
+ return len(self._cond_val)
+
+ @property
+ def cond_ext_drift(self):
+ """:class:`numpy.ndarray`: The ext. drift at the conditions."""
+ return self._cond_ext_drift
+
+ @property
+ def drift_functions(self):
+ """:class:`list` of :any:`callable`: The drift functions."""
+ return self._drift_functions
+
+ @property
+ def drift_no(self):
+ """:class:`int`: Number of drift values per point."""
+ return len(self.drift_functions) + self.cond_ext_drift.shape[0]
+
+ @property
+ def cond_trend(self):
+ """:class:`numpy.ndarray`: Trend at the conditions."""
+ return self._cond_trend
+
+ @property
+ def trend_function(self):
+ """:any:`callable`: The trend function."""
+ return self._trend_function
+
+ @trend_function.setter
+ def trend_function(self, trend_function):
+ if trend_function is None:
+ trend_function = no_trend
+ if not callable(trend_function):
+ raise ValueError("Detrended kriging: trend function not callable.")
+ self._trend_function = trend_function
+
+ @property
+ def unbiased(self):
+ """:class:`bool`: Whether the kriging is unbiased or not."""
+ return self._unbiased
+
+
+if __name__ == "__main__": # pragma: no cover
+ import doctest
+
+ doctest.testmod()
diff --git a/gstools/krige/krigesum.pyx b/gstools/krige/krigesum.pyx
index d02302b8..05a7d152 100644
--- a/gstools/krige/krigesum.pyx
+++ b/gstools/krige/krigesum.pyx
@@ -1,9 +1,8 @@
-# cython: language_level=2
+#cython: language_level=3, boundscheck=False, wraparound=False, cdivision=True
# -*- coding: utf-8 -*-
"""
This is a summator for the kriging routines
"""
-from __future__ import division, absolute_import, print_function
import numpy as np
@@ -12,9 +11,11 @@ from cython.parallel import prange
cimport numpy as np
-@cython.boundscheck(False) # turn off bounds-checking for entire function
-@cython.wraparound(False) # turn off negative index wrapping for entire function
-def krigesum(double[:,:] krig_mat, double[:,:] krig_vecs, double[:] cond):
+def krigesum(
+ const double[:,:] krig_mat,
+ const double[:,:] krig_vecs,
+ const double[:] cond
+):
cdef int mat_i = krig_mat.shape[0]
cdef int res_i = krig_vecs.shape[1]
diff --git a/gstools/krige/methods.py b/gstools/krige/methods.py
new file mode 100644
index 00000000..bad1b059
--- /dev/null
+++ b/gstools/krige/methods.py
@@ -0,0 +1,385 @@
+# -*- coding: utf-8 -*-
+"""
+GStools subpackage providing a class for simple kriging.
+
+.. currentmodule:: gstools.krige.methods
+
+The following classes are provided
+
+.. autosummary::
+ Simple
+ Ordinary
+ Universal
+ ExtDrift
+ Detrended
+"""
+# pylint: disable=C0103
+import numpy as np
+from scipy.linalg import inv
+from gstools.field.tools import make_anisotropic, rotate_mesh
+from gstools.tools.geometric import pos2xyz, xyz2pos
+from gstools.krige.base import Krige
+from gstools.krige.tools import eval_func, no_trend
+
+__all__ = ["Simple", "Ordinary", "Universal", "ExtDrift", "Detrended"]
+
+
+class Simple(Krige):
+ """
+ Simple kriging.
+
+ Simple kriging is used to interpolate data with a given mean.
+
+ Parameters
+ ----------
+ model : :any:`CovModel`
+ Covariance Model used for kriging.
+ cond_pos : :class:`list`
+ tuple, containing the given condition positions (x, [y, z])
+ cond_val : :class:`numpy.ndarray`
+ the values of the conditions
+ mean : :class:`float`, optional
+ mean value of the kriging field
+ trend_function : :any:`callable`, optional
+ A callable trend function. Should have the signiture: f(x, [y, z])
+ This is used for detrended kriging, where the trended is subtracted
+ from the conditions before kriging is applied.
+ This can be used for regression kriging, where the trend function
+ is determined by an external regression algorithm.
+ """
+
+ def __init__(
+ self, model, cond_pos, cond_val, mean=0.0, trend_function=None
+ ):
+ super().__init__(
+ model, cond_pos, cond_val, mean=mean, trend_function=trend_function
+ )
+ self._unbiased = False
+
+ def _get_krige_mat(self):
+ """Calculate the inverse matrix of the kriging equation."""
+ return inv(self.model.cov_nugget(self._get_dists(self._krige_pos)))
+
+ def _get_krige_vecs(self, pos, chunk_slice=(0, None), ext_drift=None):
+ """Calculate the RHS of the kriging equation."""
+ return self.model.cov_nugget(
+ self._get_dists(self._krige_pos, pos, chunk_slice)
+ )
+
+ def _post_field(self, field, krige_var):
+ """
+ Postprocessing and saving of kriging field and error variance.
+
+ Parameters
+ ----------
+ field : :class:`numpy.ndarray`
+ Raw kriging field.
+ krige_var : :class:`numpy.ndarray`
+ Raw kriging error variance.
+ """
+ if self.trend_function is no_trend:
+ self.field = field + self.mean
+ else:
+ self.field = (
+ field
+ + self.mean
+ + eval_func(self.trend_function, self.pos, self.mesh_type)
+ )
+ # add the given mean
+ self.krige_var = self.model.sill - krige_var
+
+ @property
+ def _krige_cond(self):
+ """:class:`numpy.ndarray`: The prepared kriging conditions."""
+ return self.cond_val - self.mean - self.cond_trend
+
+ def __repr__(self):
+ """Return String representation."""
+ return "Simple(model={0}, cond_pos={1}, cond_val={2}, mean={3})".format(
+ self.model, self.cond_pos, self.cond_val, self.mean
+ )
+
+
+class Ordinary(Krige):
+ """
+ Ordinary kriging.
+
+ Ordinary kriging is used to interpolate data and estimate a proper mean.
+
+ Parameters
+ ----------
+ model : :any:`CovModel`
+ Covariance Model used for kriging.
+ cond_pos : :class:`list`
+ tuple, containing the given condition positions (x, [y, z])
+ cond_val : :class:`numpy.ndarray`
+ the values of the conditions
+ trend_function : :any:`callable`, optional
+ A callable trend function. Should have the signiture: f(x, [y, z])
+ This is used for detrended kriging, where the trended is subtracted
+ from the conditions before kriging is applied.
+ This can be used for regression kriging, where the trend function
+ is determined by an external regression algorithm.
+ """
+
+ def __init__(self, model, cond_pos, cond_val, trend_function=None):
+ super().__init__(
+ model, cond_pos, cond_val, trend_function=trend_function
+ )
+
+ def _get_krige_mat(self):
+ """Calculate the inverse matrix of the kriging equation."""
+ size = self.cond_no + int(self.unbiased)
+ res = np.empty((size, size), dtype=np.double)
+ res[: self.cond_no, : self.cond_no] = self.model.vario_nugget(
+ self._get_dists(self._krige_pos)
+ )
+ if self.unbiased:
+ res[self.cond_no, :] = 1
+ res[:, self.cond_no] = 1
+ res[self.cond_no, self.cond_no] = 0
+ return inv(res)
+
+ def _get_krige_vecs(self, pos, chunk_slice=(0, None), ext_drift=None):
+ """Calculate the RHS of the kriging equation."""
+ chunk_size = len(pos[0]) if chunk_slice[1] is None else chunk_slice[1]
+ chunk_size -= chunk_slice[0]
+ size = self.cond_no + int(self.unbiased)
+ res = np.empty((size, chunk_size), dtype=np.double)
+ res[: self.cond_no, :] = self.model.vario_nugget(
+ self._get_dists(self._krige_pos, pos, chunk_slice)
+ )
+ if self.unbiased:
+ res[self.cond_no, :] = 1
+ return res
+
+ def get_mean(self):
+ """Calculate the estimated mean."""
+ mean_est = np.concatenate(
+ (np.full_like(self.cond_val, self.model.sill), [1])
+ )
+ return np.einsum("i,ij,j", self._krige_cond, self._krige_mat, mean_est)
+
+ def __repr__(self):
+ """Return String representation."""
+ return "Ordinary(model={0}, cond_pos={1}, cond_val={2}".format(
+ self.model, self.cond_pos, self.cond_val
+ )
+
+
+class Universal(Krige):
+ """
+ Universal kriging.
+
+ Universal kriging is used to interpolate given data with a variable mean,
+ that is determined by a functional drift.
+
+ This estimator is set to be unbiased by default.
+ This means, that the weights in the kriging equation sum up to 1.
+ Consequently no constant function needs to be given for a constant drift,
+ since the unbiased condition is applied to all given drift functions.
+
+ Parameters
+ ----------
+ model : :any:`CovModel`
+ Covariance Model used for kriging.
+ cond_pos : :class:`list`
+ tuple, containing the given condition positions (x, [y, z])
+ cond_val : :class:`numpy.ndarray`
+ the values of the conditions
+ drift_functions : :class:`list` of :any:`callable`, :class:`str` or :class:`int`
+ Either a list of callable functions, an integer representing
+ the polynomial order of the drift or one of the following strings:
+
+ * "linear" : regional linear drift (equals order=1)
+ * "quadratic" : regional quadratic drift (equals order=2)
+
+ trend_function : :any:`callable`, optional
+ A callable trend function. Should have the signiture: f(x, [y, z])
+ This is used for detrended kriging, where the trended is subtracted
+ from the conditions before kriging is applied.
+ This can be used for regression kriging, where the trend function
+ is determined by an external regression algorithm.
+ """
+
+ def __init__(
+ self, model, cond_pos, cond_val, drift_functions, trend_function=None
+ ):
+ super().__init__(
+ model,
+ cond_pos,
+ cond_val,
+ drift_functions=drift_functions,
+ trend_function=trend_function,
+ )
+
+ def _get_krige_mat(self):
+ """Calculate the inverse matrix of the kriging equation."""
+ size = self.cond_no + int(self.unbiased) + self.drift_no
+ res = np.empty((size, size), dtype=np.double)
+ res[: self.cond_no, : self.cond_no] = self.model.vario_nugget(
+ self._get_dists(self._krige_pos)
+ )
+ if self.unbiased:
+ res[self.cond_no, : self.cond_no] = 1
+ res[: self.cond_no, self.cond_no] = 1
+ for i, f in enumerate(self.drift_functions):
+ drift_tmp = f(*self.cond_pos)
+ res[-self.drift_no + i, : self.cond_no] = drift_tmp
+ res[: self.cond_no, -self.drift_no + i] = drift_tmp
+ res[self.cond_no :, self.cond_no :] = 0
+ return inv(res)
+
+ def _get_krige_vecs(self, pos, chunk_slice=(0, None), ext_drift=None):
+ """Calculate the RHS of the kriging equation."""
+ chunk_size = len(pos[0]) if chunk_slice[1] is None else chunk_slice[1]
+ chunk_size -= chunk_slice[0]
+ size = self.cond_no + int(self.unbiased) + self.drift_no
+ res = np.empty((size, chunk_size), dtype=np.double)
+ res[: self.cond_no, :] = self.model.vario_nugget(
+ self._get_dists(self._krige_pos, pos, chunk_slice)
+ )
+ if self.unbiased:
+ res[self.cond_no, :] = 1
+ # trend function need the anisotropic and rotated positions
+ if not self.model.is_isotropic:
+ x, y, z = pos2xyz(pos, max_dim=self.model.dim)
+ y, z = make_anisotropic(self.model.dim, self.model.anis, y, z)
+ if self.model.do_rotation:
+ x, y, z = rotate_mesh(
+ self.model.dim, self.model.angles, x, y, z
+ )
+ pos = xyz2pos(x, y, z, max_dim=self.model.dim)
+ chunk_pos = list(pos[: self.model.dim])
+ for i in range(self.model.dim):
+ chunk_pos[i] = chunk_pos[i][slice(*chunk_slice)]
+ for i, f in enumerate(self.drift_functions):
+ res[-self.drift_no + i, :] = f(*chunk_pos)
+ return res
+
+ def __repr__(self):
+ """Return String representation."""
+ return "Universal(model={0}, cond_pos={1}, cond_val={2})".format(
+ self.model, self.cond_pos, self.cond_val
+ )
+
+
+class ExtDrift(Krige):
+ """
+ External drift kriging (EDK).
+
+ External drift kriging is used to interpolate given data
+ with a variable mean, that is determined by an external drift.
+
+ This estimator is set to be unbiased by default.
+ This means, that the weights in the kriging equation sum up to 1.
+ Consequently no constant external drift needs to be given to estimate
+ a proper mean.
+
+ Parameters
+ ----------
+ model : :any:`CovModel`
+ Covariance Model used for kriging.
+ cond_pos : :class:`list`
+ tuple, containing the given condition positions (x, [y, z])
+ cond_val : :class:`numpy.ndarray`
+ the values of the conditions
+ ext_drift : :class:`numpy.ndarray`
+ the external drift values at the given condition positions.
+ trend_function : :any:`callable`, optional
+ A callable trend function. Should have the signiture: f(x, [y, z])
+ This is used for detrended kriging, where the trended is subtracted
+ from the conditions before kriging is applied.
+ This can be used for regression kriging, where the trend function
+ is determined by an external regression algorithm.
+ """
+
+ def __init__(
+ self, model, cond_pos, cond_val, ext_drift, trend_function=None
+ ):
+ super().__init__(
+ model,
+ cond_pos,
+ cond_val,
+ ext_drift=ext_drift,
+ trend_function=trend_function,
+ )
+
+ def _get_krige_mat(self):
+ """Calculate the inverse matrix of the kriging equation."""
+ size = self.cond_no + int(self.unbiased) + self.drift_no
+ res = np.empty((size, size), dtype=np.double)
+ res[: self.cond_no, : self.cond_no] = self.model.vario_nugget(
+ self._get_dists(self._krige_pos)
+ )
+ if self.unbiased:
+ res[self.cond_no, : self.cond_no] = 1
+ res[: self.cond_no, self.cond_no] = 1
+ res[-self.drift_no :, : self.cond_no] = self.cond_ext_drift
+ res[: self.cond_no, -self.drift_no :] = self.cond_ext_drift.T
+ res[self.cond_no :, self.cond_no :] = 0
+ return inv(res)
+
+ def _get_krige_vecs(self, pos, chunk_slice=(0, None), ext_drift=None):
+ """Calculate the RHS of the kriging equation."""
+ chunk_size = len(pos[0]) if chunk_slice[1] is None else chunk_slice[1]
+ chunk_size -= chunk_slice[0]
+ size = self.cond_no + int(self.unbiased) + self.drift_no
+ res = np.empty((size, chunk_size), dtype=np.double)
+ res[: self.cond_no, :] = self.model.vario_nugget(
+ self._get_dists(self._krige_pos, pos, chunk_slice)
+ )
+ if self.unbiased:
+ res[self.cond_no, :] = 1
+ res[-self.drift_no :, :] = ext_drift[:, slice(*chunk_slice)]
+ return res
+
+ def __repr__(self):
+ """Return String representation."""
+ return "ExtDrift(model={0}, cond_pos={1}, cond_val={2})".format(
+ self.model, self.cond_pos, self.cond_val
+ )
+
+
+class Detrended(Simple):
+ """
+ Detrended simple kriging.
+
+ In detrended kriging, the data is detrended before interpolation by
+ simple kriging with zero mean.
+
+ The trend needs to be a callable function the user has to provide.
+ This can be used for regression kriging, where the trend function
+ is determined by an external regression algorithm.
+
+ This is just a shortcut for simple kriging with a given trend function
+ and zero mean. A trend can be given with EVERY provided kriging routine.
+
+ Parameters
+ ----------
+ model : :any:`CovModel`
+ Covariance Model used for kriging.
+ cond_pos : :class:`list`
+ tuple, containing the given condition positions (x, [y, z])
+ cond_val : :class:`numpy.ndarray`
+ the values of the conditions
+ trend_function : :any:`callable`
+ The callable trend function. Should have the signiture: f(x, [y, z])
+ """
+
+ def __init__(self, model, cond_pos, cond_val, trend_function):
+ super().__init__(
+ model, cond_pos, cond_val, trend_function=trend_function
+ )
+
+ def __repr__(self):
+ """Return String representation."""
+ return "Detrended(model={0} cond_pos={1}, cond_val={2})".format(
+ self.model, self.cond_pos, self.cond_val
+ )
+
+
+if __name__ == "__main__": # pragma: no cover
+ import doctest
+
+ doctest.testmod()
diff --git a/gstools/krige/ordinary.py b/gstools/krige/ordinary.py
deleted file mode 100644
index cd8a6b94..00000000
--- a/gstools/krige/ordinary.py
+++ /dev/null
@@ -1,183 +0,0 @@
-# -*- coding: utf-8 -*-
-"""
-GStools subpackage providing a class for ordinary kriging.
-
-.. currentmodule:: gstools.krige.ordinary
-
-The following classes are provided
-
-.. autosummary::
- Ordinary
-"""
-# pylint: disable=C0103
-from __future__ import division, absolute_import, print_function
-
-import numpy as np
-from scipy.linalg import inv
-from scipy.spatial.distance import cdist
-
-from gstools.field.tools import (
- check_mesh,
- make_isotropic,
- unrotate_mesh,
- reshape_axis_from_struct_to_unstruct,
- reshape_field_from_unstruct_to_struct,
-)
-from gstools.field.base import Field
-from gstools.tools.geometric import pos2xyz, xyz2pos
-from gstools.krige.krigesum import krigesum
-from gstools.krige.tools import set_condition
-
-__all__ = ["Ordinary"]
-
-
-class Ordinary(Field):
- """
- A class for ordinary kriging.
-
- Parameters
- ----------
- model : :any:`CovModel`
- Covariance Model used for kriging.
- cond_pos : :class:`list`
- tuple, containing the given condition positions (x, [y, z])
- cond_val : :class:`numpy.ndarray`
- the values of the conditions
- """
-
- def __init__(self, model, cond_pos, cond_val):
- super(Ordinary, self).__init__(model, mean=0.0)
- self.krige_var = None
- # initialize private attributes
- self._value_type = "scalar"
- self._cond_pos = None
- self._cond_val = None
- self.set_condition(cond_pos, cond_val)
-
- def __call__(self, pos, mesh_type="unstructured"):
- """
- Generate the ordinary kriging field.
-
- The field is saved as `self.field` and is also returned.
-
- Parameters
- ----------
- pos : :class:`list`
- the position tuple, containing main direction and transversal
- directions (x, [y, z])
- mesh_type : :class:`str`
- 'structured' / 'unstructured'
-
- Returns
- -------
- field : :class:`numpy.ndarray`
- the kriged field
- krige_var : :class:`numpy.ndarray`
- the kriging error variance
- """
- # internal conversation
- x, y, z = pos2xyz(pos, dtype=np.double, max_dim=self.model.dim)
- c_x, c_y, c_z = pos2xyz(
- self.cond_pos, dtype=np.double, max_dim=self.model.dim
- )
- self.pos = xyz2pos(x, y, z)
- self.mesh_type = mesh_type
- # format the positional arguments of the mesh
- check_mesh(self.model.dim, x, y, z, mesh_type)
- mesh_type_changed = False
- if mesh_type == "structured":
- mesh_type_changed = True
- mesh_type_old = mesh_type
- mesh_type = "unstructured"
- x, y, z, axis_lens = reshape_axis_from_struct_to_unstruct(
- self.model.dim, x, y, z
- )
- if self.model.do_rotation:
- x, y, z = unrotate_mesh(self.model.dim, self.model.angles, x, y, z)
- c_x, c_y, c_z = unrotate_mesh(
- self.model.dim, self.model.angles, c_x, c_y, c_z
- )
- y, z = make_isotropic(self.model.dim, self.model.anis, y, z)
- c_y, c_z = make_isotropic(self.model.dim, self.model.anis, c_y, c_z)
-
- # set condtions
- cond = np.concatenate((self.cond_val, [0]))
- krig_mat = inv(self._get_krig_mat((c_x, c_y, c_z), (c_x, c_y, c_z)))
- krig_vecs = self._get_vario_mat((c_x, c_y, c_z), (x, y, z), add=True)
- # generate the kriged field
- field, krige_var = krigesum(krig_mat, krig_vecs, cond)
- # calculate the estimated mean (kriging field at infinity)
- mean_est = np.concatenate(
- (np.full_like(self.cond_val, self.model.sill), [1])
- )
- self.mean = np.einsum("i,ij,j", cond, krig_mat, mean_est)
-
- # reshape field if we got an unstructured mesh
- if mesh_type_changed:
- mesh_type = mesh_type_old
- field = reshape_field_from_unstruct_to_struct(
- self.model.dim, field, axis_lens
- )
- krige_var = reshape_field_from_unstruct_to_struct(
- self.model.dim, krige_var, axis_lens
- )
- # save the field
- self.krige_var = krige_var
- self.field = field
- return self.field, self.krige_var
-
- def _get_krig_mat(self, pos1, pos2):
- size = pos1[0].size
- res = np.empty((size + 1, size + 1), dtype=np.double)
- res[:size, :size] = self._get_vario_mat(pos1, pos2)
- res[size, :] = 1
- res[:, size] = 1
- res[size, size] = 0
- return res
-
- def _get_vario_mat(self, pos1, pos2, add=False):
- res = self.model.vario_nugget(
- cdist(
- np.column_stack(pos1[: self.model.dim]),
- np.column_stack(pos2[: self.model.dim]),
- )
- )
- if add:
- return np.vstack((res, np.ones((1, res.shape[1]))))
- return res
-
- def set_condition(self, cond_pos, cond_val):
- """Set the conditions for kriging.
-
- Parameters
- ----------
- cond_pos : :class:`list`
- the position tuple of the conditions (x, [y, z])
- cond_val : :class:`numpy.ndarray`
- the values of the conditions
- """
- self._cond_pos, self._cond_val = set_condition(
- cond_pos, cond_val, self.model.dim
- )
-
- @property
- def cond_pos(self):
- """:class:`list`: The position tuple of the conditions."""
- return self._cond_pos
-
- @property
- def cond_val(self):
- """:class:`list`: The values of the conditions."""
- return self._cond_val
-
- def __repr__(self):
- """Return String representation."""
- return "Ordinary(model={0}, cond_pos={1}, cond_val={2}".format(
- self.model, self.cond_pos, self.cond_val
- )
-
-
-if __name__ == "__main__": # pragma: no cover
- import doctest
-
- doctest.testmod()
diff --git a/gstools/krige/simple.py b/gstools/krige/simple.py
deleted file mode 100644
index ca1ba7f5..00000000
--- a/gstools/krige/simple.py
+++ /dev/null
@@ -1,170 +0,0 @@
-# -*- coding: utf-8 -*-
-"""
-GStools subpackage providing a class for simple kriging.
-
-.. currentmodule:: gstools.krige.simple
-
-The following classes are provided
-
-.. autosummary::
- Simple
-"""
-# pylint: disable=C0103
-from __future__ import division, absolute_import, print_function
-
-
-import numpy as np
-from scipy.linalg import inv
-from scipy.spatial.distance import cdist
-
-from gstools.field.tools import (
- check_mesh,
- make_isotropic,
- unrotate_mesh,
- reshape_axis_from_struct_to_unstruct,
- reshape_field_from_unstruct_to_struct,
-)
-from gstools.field.base import Field
-from gstools.tools.geometric import pos2xyz, xyz2pos
-from gstools.krige.krigesum import krigesum
-from gstools.krige.tools import set_condition
-
-__all__ = ["Simple"]
-
-
-class Simple(Field):
- """
- A class for simple kriging.
-
- Parameters
- ----------
- model : :any:`CovModel`
- Covariance Model used for kriging.
- mean : :class:`float`, optional
- mean value of the kriging field
- cond_pos : :class:`list`
- tuple, containing the given condition positions (x, [y, z])
- cond_val : :class:`numpy.ndarray`
- the values of the conditions
- """
-
- def __init__(self, model, mean, cond_pos, cond_val):
- super(Simple, self).__init__(model, mean)
- self.krige_var = None
- # initialize private attributes
- self._value_type = "scalar"
- self._cond_pos = None
- self._cond_val = None
- self.set_condition(cond_pos, cond_val)
-
- def __call__(self, pos, mesh_type="unstructured"):
- """
- Generate the simple kriging field.
-
- The field is saved as `self.field` and is also returned.
-
- Parameters
- ----------
- pos : :class:`list`
- the position tuple, containing main direction and transversal
- directions (x, [y, z])
- mesh_type : :class:`str`
- 'structured' / 'unstructured'
-
- Returns
- -------
- field : :class:`numpy.ndarray`
- the kriged field
- krige_var : :class:`numpy.ndarray`
- the kriging error variance
- """
- # internal conversation
- x, y, z = pos2xyz(pos, dtype=np.double, max_dim=self.model.dim)
- c_x, c_y, c_z = pos2xyz(
- self.cond_pos, dtype=np.double, max_dim=self.model.dim
- )
- self.pos = xyz2pos(x, y, z)
- self.mesh_type = mesh_type
- # format the positional arguments of the mesh
- check_mesh(self.model.dim, x, y, z, mesh_type)
- mesh_type_changed = False
- if mesh_type == "structured":
- mesh_type_changed = True
- mesh_type_old = mesh_type
- mesh_type = "unstructured"
- x, y, z, axis_lens = reshape_axis_from_struct_to_unstruct(
- self.model.dim, x, y, z
- )
- if self.model.do_rotation:
- x, y, z = unrotate_mesh(self.model.dim, self.model.angles, x, y, z)
- c_x, c_y, c_z = unrotate_mesh(
- self.model.dim, self.model.angles, c_x, c_y, c_z
- )
- y, z = make_isotropic(self.model.dim, self.model.anis, y, z)
- c_y, c_z = make_isotropic(self.model.dim, self.model.anis, c_y, c_z)
-
- # set condtions to zero mean
- cond = self.cond_val - self.mean
- krig_mat = inv(self._get_cov_mat((c_x, c_y, c_z), (c_x, c_y, c_z)))
- krig_vecs = self._get_cov_mat((c_x, c_y, c_z), (x, y, z))
- # generate the kriged field
- field, krige_var = krigesum(krig_mat, krig_vecs, cond)
-
- # reshape field if we got an unstructured mesh
- if mesh_type_changed:
- mesh_type = mesh_type_old
- field = reshape_field_from_unstruct_to_struct(
- self.model.dim, field, axis_lens
- )
- krige_var = reshape_field_from_unstruct_to_struct(
- self.model.dim, krige_var, axis_lens
- )
- # calculate the kriging error
- self.krige_var = self.model.sill - krige_var
- # add the given mean
- self.field = field + self.mean
- return self.field, self.krige_var
-
- def _get_cov_mat(self, pos1, pos2):
- return self.model.cov_nugget(
- cdist(
- np.column_stack(pos1[: self.model.dim]),
- np.column_stack(pos2[: self.model.dim]),
- )
- )
-
- def set_condition(self, cond_pos, cond_val):
- """Set the conditions for kriging.
-
- Parameters
- ----------
- cond_pos : :class:`list`
- the position tuple of the conditions (x, [y, z])
- cond_val : :class:`numpy.ndarray`
- the values of the conditions
- """
- self._cond_pos, self._cond_val = set_condition(
- cond_pos, cond_val, self.model.dim
- )
-
- @property
- def cond_pos(self):
- """:class:`list`: The position tuple of the conditions."""
- return self._cond_pos
-
- @property
- def cond_val(self):
- """:class:`list`: The values of the conditions."""
- return self._cond_val
-
- def __repr__(self):
- """Return String representation."""
- return "Simple(model={0}, mean={1}, cond_pos={2}, cond_val={3}".format(
- self.model, self.mean, self.cond_pos, self.cond_val
- )
-
-
-if __name__ == "__main__": # pragma: no cover
- import doctest
-
- doctest.testmod()
diff --git a/gstools/krige/tools.py b/gstools/krige/tools.py
index d316269b..121e312b 100644
--- a/gstools/krige/tools.py
+++ b/gstools/krige/tools.py
@@ -8,18 +8,73 @@
.. autosummary::
set_condition
+ get_drift_functions
+ no_trend
+ eval_func
"""
# pylint: disable=C0103
-from __future__ import print_function, division, absolute_import
-
+from itertools import combinations_with_replacement
import numpy as np
from gstools.tools.geometric import pos2xyz, xyz2pos
+from gstools.field.tools import (
+ reshape_axis_from_struct_to_unstruct,
+ reshape_field_from_unstruct_to_struct,
+)
+
+__all__ = ["no_trend", "eval_func", "set_condition", "get_drift_functions"]
+
+
+def no_trend(*args, **kwargs):
+ """
+ Zero trend dummy function.
+
+ Parameters
+ ----------
+ *args : any
+ Ignored arguments.
+ **kwargs : any
+ Ignored keyword arguments.
+
+ Returns
+ -------
+ float
+ A zero trend given as single float.
+
+ """
+ return 0.0
-__all__ = ["set_condition"]
+
+def eval_func(func, pos, mesh_type="structured"):
+ """
+ Evaluate a function on a mesh.
+
+ Parameters
+ ----------
+ func : :any:`callable`
+ The function to be called. Should have the signiture f(x, [y, z])
+ pos : :class:`list`
+ the position tuple, containing main direction and transversal
+ directions (x, [y, z])
+ mesh_type : :class:`str`, optional
+ 'structured' / 'unstructured'
+
+ Returns
+ -------
+ :class:`numpy.ndarray`
+ Function values at the given points.
+ """
+ x, y, z, dim = pos2xyz(pos, calc_dim=True)
+ if mesh_type == "structured":
+ x, y, z, axis_lens = reshape_axis_from_struct_to_unstruct(dim, x, y, z)
+ res = func(*[x, y, z][:dim])
+ if mesh_type == "structured":
+ res = reshape_field_from_unstruct_to_struct(dim, res, axis_lens)
+ return res
def set_condition(cond_pos, cond_val, max_dim=3):
- """Set the conditions for kriging.
+ """
+ Set the conditions for kriging.
Parameters
----------
@@ -30,6 +85,11 @@ def set_condition(cond_pos, cond_val, max_dim=3):
max_dim : :class:`int`, optional
Cut of information above the given dimension. Default: 3
+ Raises
+ ------
+ ValueError
+ If the given data does not match the given dimension.
+
Returns
-------
cond_pos : :class:`list`
@@ -40,6 +100,11 @@ def set_condition(cond_pos, cond_val, max_dim=3):
# convert the input for right shapes and dimension checks
c_x, c_y, c_z = pos2xyz(cond_pos, dtype=np.double, max_dim=max_dim)
cond_pos = xyz2pos(c_x, c_y, c_z)
+ if len(cond_pos) != max_dim:
+ raise ValueError(
+ "Please check your 'cond_pos' parameters. "
+ + "The dimension does not match with the given one."
+ )
cond_val = np.array(cond_val, dtype=np.double).reshape(-1)
if not all([len(cond_pos[i]) == len(cond_val) for i in range(max_dim)]):
raise ValueError(
@@ -47,3 +112,44 @@ def set_condition(cond_pos, cond_val, max_dim=3):
+ "The shapes do not match."
)
return cond_pos, cond_val
+
+
+def get_drift_functions(dim, drift_type):
+ """
+ Get functions for a given drift type in universal kriging.
+
+ Parameters
+ ----------
+ dim : :class:`int`
+ Given dimension.
+ drift_type : :class:`str` or :class:`int`
+ Drift type: 'linear' or 'quadratic' or an integer for the polynomial
+ order of the drift type. (linear equals 1, quadratic equals 2 ...)
+
+ Returns
+ -------
+ :class:`list` of :any:`callable`
+ List of drift functions.
+ """
+ if drift_type in ["lin", "linear"]:
+ drift_type = 1
+ elif drift_type in ["quad", "quadratic"]:
+ drift_type = 2
+ else:
+ drift_type = int(drift_type)
+ drift_functions = []
+ for d in range(drift_type):
+ selects = combinations_with_replacement(range(dim), d + 1)
+ for select in selects:
+ drift_functions.append(_f_factory(select))
+ return drift_functions
+
+
+def _f_factory(select):
+ def f(*pos):
+ res = 1.0
+ for i in select:
+ res *= np.asarray(pos[i])
+ return res
+
+ return f
diff --git a/gstools/random/__init__.py b/gstools/random/__init__.py
index 375a9b55..ec78b43c 100644
--- a/gstools/random/__init__.py
+++ b/gstools/random/__init__.py
@@ -24,7 +24,6 @@
----
"""
-from __future__ import absolute_import
from gstools.random.rng import RNG
from gstools.random.tools import MasterRNG, dist_gen
diff --git a/gstools/random/rng.py b/gstools/random/rng.py
index 9b37f8c9..e2915f6f 100644
--- a/gstools/random/rng.py
+++ b/gstools/random/rng.py
@@ -10,7 +10,6 @@
RNG
"""
# pylint: disable=no-member
-from __future__ import division, absolute_import, print_function
import numpy as np
import numpy.random as rand
@@ -21,7 +20,7 @@
__all__ = ["RNG"]
-class RNG(object):
+class RNG:
"""
A random number generator for different distributions and multiple streams.
diff --git a/gstools/random/tools.py b/gstools/random/tools.py
index 6f445e7c..c4799ee4 100644
--- a/gstools/random/tools.py
+++ b/gstools/random/tools.py
@@ -10,7 +10,6 @@
MasterRNG
dist_gen
"""
-from __future__ import division, absolute_import, print_function
from scipy.stats import rv_continuous
import numpy.random as rand
@@ -18,7 +17,7 @@
__all__ = ["MasterRNG", "dist_gen"]
-class MasterRNG(object):
+class MasterRNG:
"""Master random number generator for generating seeds.
Parameters
@@ -108,7 +107,7 @@ class DistPdf(rv_continuous):
def __init__(self, pdf_in, **kwargs):
self.pdf_in = pdf_in
- super(DistPdf, self).__init__(**kwargs)
+ super().__init__(**kwargs)
def _pdf(self, x, *args):
return self.pdf_in(x)
@@ -119,7 +118,7 @@ class DistCdf(rv_continuous):
def __init__(self, cdf_in, **kwargs):
self.cdf_in = cdf_in
- super(DistCdf, self).__init__(**kwargs)
+ super().__init__(**kwargs)
def _cdf(self, x, *args):
return self.cdf_in(x)
@@ -131,7 +130,7 @@ class DistPdfCdf(rv_continuous):
def __init__(self, pdf_in, cdf_in, **kwargs):
self.pdf_in = pdf_in
self.cdf_in = cdf_in
- super(DistPdfCdf, self).__init__(**kwargs)
+ super().__init__(**kwargs)
def _pdf(self, x, *args):
return self.pdf_in(x)
@@ -146,7 +145,7 @@ class DistPdfPpf(rv_continuous):
def __init__(self, pdf_in, ppf_in, **kwargs):
self.pdf_in = pdf_in
self.ppf_in = ppf_in
- super(DistPdfPpf, self).__init__(**kwargs)
+ super().__init__(**kwargs)
def _pdf(self, x, *args):
return self.pdf_in(x)
@@ -161,7 +160,7 @@ class DistCdfPpf(rv_continuous):
def __init__(self, cdf_in, ppf_in, **kwargs):
self.cdf_in = cdf_in
self.ppf_in = ppf_in
- super(DistCdfPpf, self).__init__(**kwargs)
+ super().__init__(**kwargs)
def _cdf(self, x, *args):
return self.cdf_in(x)
@@ -177,7 +176,7 @@ def __init__(self, pdf_in, cdf_in, ppf_in, **kwargs):
self.pdf_in = pdf_in
self.cdf_in = cdf_in
self.ppf_in = ppf_in
- super(DistPdfCdfPpf, self).__init__(**kwargs)
+ super().__init__(**kwargs)
def _pdf(self, x, *args):
return self.pdf_in(x)
diff --git a/gstools/tools/__init__.py b/gstools/tools/__init__.py
index 3f430c9a..12de7d00 100644
--- a/gstools/tools/__init__.py
+++ b/gstools/tools/__init__.py
@@ -8,12 +8,12 @@
^^^^^^
.. autosummary::
- to_vtk_structured
+ vtk_export
vtk_export_structured
- to_vtk_unstructured
vtk_export_unstructured
to_vtk
- vtk_export
+ to_vtk_structured
+ to_vtk_unstructured
Special functions
^^^^^^^^^^^^^^^^^
@@ -23,6 +23,8 @@
exp_int
inc_beta
tplstable_cor
+ tpl_exp_spec_dens
+ tpl_gau_spec_dens
Geometric
^^^^^^^^^
@@ -36,26 +38,40 @@
----
"""
-from __future__ import absolute_import
from gstools.tools.export import (
+ to_vtk,
+ to_vtk_structured,
+ to_vtk_unstructured,
vtk_export_structured,
vtk_export_unstructured,
vtk_export,
)
-from gstools.tools.special import inc_gamma, exp_int, inc_beta, tplstable_cor
+from gstools.tools.special import (
+ inc_gamma,
+ exp_int,
+ inc_beta,
+ tplstable_cor,
+ tpl_exp_spec_dens,
+ tpl_gau_spec_dens,
+)
from gstools.tools.geometric import r3d_x, r3d_y, r3d_z, xyz2pos, pos2xyz
__all__ = [
+ "vtk_export",
"vtk_export_structured",
"vtk_export_unstructured",
- "vtk_export",
+ "to_vtk",
+ "to_vtk_structured",
+ "to_vtk_unstructured",
"inc_gamma",
"exp_int",
"inc_beta",
"tplstable_cor",
+ "tpl_exp_spec_dens",
+ "tpl_gau_spec_dens",
"xyz2pos",
"pos2xyz",
"r3d_x",
diff --git a/gstools/tools/export.py b/gstools/tools/export.py
index 65fc9989..9174830b 100644
--- a/gstools/tools/export.py
+++ b/gstools/tools/export.py
@@ -7,15 +7,14 @@
The following functions are provided
.. autosummary::
- to_vtk_structured
+ vtk_export
vtk_export_structured
- to_vtk_unstructured
vtk_export_unstructured
to_vtk
- vtk_export
+ to_vtk_structured
+ to_vtk_unstructured
"""
# pylint: disable=C0103, E1101
-from __future__ import print_function, division, absolute_import
import numpy as np
from pyevtk.hl import gridToVTK, pointsToVTK
diff --git a/gstools/tools/geometric.py b/gstools/tools/geometric.py
index de297a4e..6edcd758 100644
--- a/gstools/tools/geometric.py
+++ b/gstools/tools/geometric.py
@@ -14,7 +14,6 @@
xyz2pos
"""
# pylint: disable=C0103
-from __future__ import print_function, division, absolute_import
import numpy as np
@@ -150,7 +149,7 @@ def xyz2pos(x, y=None, z=None, dtype=None, max_dim=3):
Returns
-------
- pos : :class:`numpy.ndarray`
+ pos : :class:`tuple` of :class:`numpy.ndarray`
the position tuple
"""
if y is None and z is not None:
diff --git a/gstools/tools/special.py b/gstools/tools/special.py
index 5100fae9..27241717 100644
--- a/gstools/tools/special.py
+++ b/gstools/tools/special.py
@@ -11,14 +11,22 @@
exp_int
inc_beta
tplstable_cor
+ tpl_exp_spec_dens
+ tpl_gau_spec_dens
"""
# pylint: disable=C0103, E1101
-from __future__ import print_function, division, absolute_import
import numpy as np
from scipy import special as sps
-__all__ = ["inc_gamma", "exp_int", "inc_beta", "tplstable_cor"]
+__all__ = [
+ "inc_gamma",
+ "exp_int",
+ "inc_beta",
+ "tplstable_cor",
+ "tpl_exp_spec_dens",
+ "tpl_gau_spec_dens",
+]
# special functions ###########################################################
@@ -81,13 +89,30 @@ def exp_int(s, x):
return res
+def inc_beta(a, b, x):
+ r"""The incomplete Beta function.
+
+ Given by: :math:`B(a,b;\,x) = \int_0^x t^{a-1}\,(1-t)^{b-1}\,dt`
+
+ Parameters
+ ----------
+ a : :class:`float`
+ first exponent in the integral
+ b : :class:`float`
+ second exponent in the integral
+ x : :class:`numpy.ndarray`
+ input values
+ """
+ return sps.betainc(a, b, x) * sps.beta(a, b)
+
+
def tplstable_cor(r, len_scale, hurst, alpha):
r"""The correlation function of the TPLStable model.
- Given by
+ Given by the following correlation function:
.. math::
- \mathrm{cor}(r) =
+ \rho(r) =
\frac{2H}{\alpha} \cdot
E_{1+\frac{2H}{\alpha}}
\left(\left(\frac{r}{\ell}\right)^{\alpha} \right)
@@ -113,18 +138,82 @@ def tplstable_cor(r, len_scale, hurst, alpha):
return res
-def inc_beta(a, b, x):
- r"""The incomplete Beta function.
+def tpl_exp_spec_dens(k, dim, len_scale, hurst, len_low=0.0):
+ r"""
+ Spectal density of the TPLExponential covariance model.
- Given by: :math:`B(a,b;\,x) = \int_0^x t^{a-1}\,(1-t)^{b-1}\,dt`
+ Parameters
+ ----------
+ k : :class:`float`
+ Radius of the phase: :math:`k=\left\Vert\mathbf{k}\right\Vert`
+ dim : :class:`int`
+ Dimension of the model.
+ len_scale : :class:`float`
+ Length scale of the model.
+ hurst : :class:`float`
+ Hurst coefficient of the power law.
+ len_low : :class:`float`, optional
+ The lower length scale truncation of the model.
+ Default: 0.0
+
+ Returns
+ -------
+ :class:`float`
+ spectal density of the TPLExponential model
+ """
+ if np.isclose(len_low, 0.0):
+ k = np.array(k, dtype=np.double)
+ z = (k * len_scale) ** 2
+ a = hurst + dim / 2.0
+ b = hurst + 0.5
+ c = hurst + dim / 2.0 + 1.0
+ d = dim / 2.0 + 0.5
+ fac = len_scale ** dim * hurst * sps.gamma(d) / (np.pi ** d * a)
+ return fac / (1.0 + z) ** a * sps.hyp2f1(a, b, c, z / (1.0 + z))
+ fac_up = (len_scale + len_low) ** (2 * hurst)
+ spec_up = tpl_exp_spec_dens(k, dim, len_scale + len_low, hurst)
+ fac_low = len_low ** (2 * hurst)
+ spec_low = tpl_exp_spec_dens(k, dim, len_low, hurst)
+ return (fac_up * spec_up - fac_low * spec_low) / (fac_up - fac_low)
+
+
+def tpl_gau_spec_dens(k, dim, len_scale, hurst, len_low=0.0):
+ r"""
+ Spectal density of the TPLGaussian covariance model.
Parameters
----------
- a : :class:`float`
- first exponent in the integral
- b : :class:`float`
- second exponent in the integral
- x : :class:`numpy.ndarray`
- input values
+ k : :class:`float`
+ Radius of the phase: :math:`k=\left\Vert\mathbf{k}\right\Vert`
+ dim : :class:`int`
+ Dimension of the model.
+ len_scale : :class:`float`
+ Length scale of the model.
+ hurst : :class:`float`
+ Hurst coefficient of the power law.
+ len_low : :class:`float`, optional
+ The lower length scale truncation of the model.
+ Default: 0.0
+
+ Returns
+ -------
+ :class:`float`
+ spectal density of the TPLExponential model
"""
- return sps.betainc(a, b, x) * sps.beta(a, b)
+ if np.isclose(len_low, 0.0):
+ k = np.array(k, dtype=np.double)
+ z = np.array((k * len_scale / 2.0) ** 2)
+ res = np.empty_like(z)
+ z_gz = z > 0.1 # greater zero
+ z_nz = np.logical_not(z_gz) # near zero
+ a = hurst + dim / 2.0
+ fac = (len_scale / 2.0) ** dim * hurst / np.pi ** (dim / 2.0)
+ res[z_gz] = fac * (sps.gamma(a) - inc_gamma(a, z[z_gz])) / z[z_gz] ** a
+ # first order approximation for z near zero
+ res[z_nz] = fac * (1.0 / a - z[z_nz] / (a + 1.0))
+ return res
+ fac_up = (len_scale + len_low) ** (2 * hurst)
+ spec_up = tpl_gau_spec_dens(k, dim, len_scale + len_low, hurst)
+ fac_low = len_low ** (2 * hurst)
+ spec_low = tpl_gau_spec_dens(k, dim, len_low, hurst)
+ return (fac_up * spec_up - fac_low * spec_low) / (fac_up - fac_low)
diff --git a/gstools/transform/__init__.py b/gstools/transform/__init__.py
index b54db482..1937e6a7 100644
--- a/gstools/transform/__init__.py
+++ b/gstools/transform/__init__.py
@@ -9,6 +9,7 @@
.. autosummary::
binary
+ discrete
boxcox
zinnharvey
normal_force_moments
@@ -19,10 +20,10 @@
----
"""
-from __future__ import absolute_import
from gstools.transform.field import (
binary,
+ discrete,
boxcox,
zinnharvey,
normal_force_moments,
@@ -34,6 +35,7 @@
__all__ = [
"binary",
+ "discrete",
"boxcox",
"zinnharvey",
"normal_force_moments",
diff --git a/gstools/transform/field.py b/gstools/transform/field.py
index 0d0b4b3e..eb0a1954 100644
--- a/gstools/transform/field.py
+++ b/gstools/transform/field.py
@@ -8,6 +8,7 @@
.. autosummary::
binary
+ discrete
boxcox
zinnharvey
normal_force_moments
@@ -17,7 +18,6 @@
normal_to_uquad
"""
# pylint: disable=C0103, E1101
-from __future__ import print_function, division, absolute_import
from warnings import warn
@@ -27,6 +27,7 @@
__all__ = [
"binary",
+ "discrete",
"boxcox",
"zinnharvey",
"normal_force_moments",
@@ -64,8 +65,72 @@ def binary(fld, divide=None, upper=None, lower=None):
divide = fld.mean if divide is None else divide
upper = fld.mean + np.sqrt(fld.model.sill) if upper is None else upper
lower = fld.mean - np.sqrt(fld.model.sill) if lower is None else lower
- fld.field[fld.field > divide] = upper
- fld.field[fld.field <= divide] = lower
+ discrete(fld, [lower, upper], thresholds=[divide])
+
+
+def discrete(fld, values, thresholds="arithmetic"):
+ """
+ Discrete transformation.
+
+ After this transformation, the field has only `len(values)` discrete
+ values.
+
+ Parameters
+ ----------
+ fld : :any:`Field`
+ Spatial Random Field class containing a generated field.
+ Field will be transformed inplace.
+ values : :any:`np.ndarray`
+ The discrete values the field will take
+ thresholds : :class:`str` or :any:`np.ndarray`, optional
+ the thresholds, where the value classes are separated
+ possible values are:
+ * "arithmetic": the mean of the 2 neighbouring values
+ * "equal": devide the field into equal parts
+ * an array of explicitly given thresholds
+ Default: "arithmetic"
+ """
+ if fld.field is None:
+ print("discrete: no field stored in SRF class.")
+ else:
+ if thresholds == "arithmetic":
+ # just in case, sort the values
+ values = np.sort(values)
+ thresholds = (values[1:] + values[:-1]) / 2
+ elif thresholds == "equal":
+ values = np.array(values)
+ n = len(values)
+ p = np.arange(1, n) / n # n-1 equal subdivisions of [0, 1]
+ rescale = np.sqrt(fld.model.sill * 2)
+ # use quantile of the normal distribution to get equal ratios
+ thresholds = fld.mean + rescale * erfinv(2 * p - 1)
+ else:
+ if len(values) != len(thresholds) + 1:
+ raise ValueError(
+ "discrete transformation: "
+ + "len(values) != len(thresholds) + 1"
+ )
+ values = np.array(values)
+ thresholds = np.array(thresholds)
+ # check thresholds
+ if not np.all(thresholds[:-1] < thresholds[1:]):
+ raise ValueError(
+ "discrete transformation: "
+ + "thresholds need to be ascending."
+ )
+ # use a separate result so the intermediate results are not affected
+ result = np.empty_like(fld.field)
+ # handle edge cases
+ result[fld.field <= thresholds[0]] = values[0]
+ result[fld.field > thresholds[-1]] = values[-1]
+ for i, value in enumerate(values[1:-1]):
+ result[
+ np.logical_and(
+ thresholds[i] < fld.field, fld.field <= thresholds[i + 1]
+ )
+ ] = value
+ # overwrite the field
+ fld.field = result
def boxcox(fld, lmbda=1, shift=0):
@@ -270,12 +335,12 @@ def _zinnharvey(field, conn="high", mean=None, var=None):
mean = np.mean(field)
if var is None:
var = np.var(field)
- field = np.abs((field - mean) / var)
+ field = np.abs((field - mean) / np.sqrt(var))
field = 2 * erf(field / np.sqrt(2)) - 1
field = np.sqrt(2) * erfinv(field)
if conn == "high":
field = -field
- return field * var + mean
+ return field * np.sqrt(var) + mean
def _normal_force_moments(field, mean=0, var=1):
@@ -443,5 +508,5 @@ def _uniform_to_uquad(field, a=0, b=1):
y_raw = 3 * field / al + ga
out = np.zeros_like(y_raw)
out[y_raw > 0] = y_raw[y_raw > 0] ** (1 / 3)
- out[y_raw < 0] = -(-y_raw[y_raw < 0]) ** (1 / 3)
+ out[y_raw < 0] = -((-y_raw[y_raw < 0]) ** (1 / 3))
return out + be
diff --git a/gstools/variogram/__init__.py b/gstools/variogram/__init__.py
index 264888a7..c5735c52 100644
--- a/gstools/variogram/__init__.py
+++ b/gstools/variogram/__init__.py
@@ -13,7 +13,6 @@
----
"""
-from __future__ import absolute_import
from gstools.variogram.variogram import (
vario_estimate_structured,
diff --git a/gstools/variogram/estimator.pyx b/gstools/variogram/estimator.pyx
index 0fe8739f..8762b946 100644
--- a/gstools/variogram/estimator.pyx
+++ b/gstools/variogram/estimator.pyx
@@ -1,18 +1,16 @@
-#!python
-# cython: language_level=2
+#cython: language_level=3, boundscheck=False, wraparound=False, cdivision=True
+# distutils: language = c++
# -*- coding: utf-8 -*-
"""
This is the variogram estimater, implemented in cython.
"""
-#!python
-#cython: language_level=2
-from __future__ import division, absolute_import, print_function
import numpy as np
cimport cython
-from cython.parallel import prange
-from libc.math cimport sqrt
+from cython.parallel import prange, parallel
+from libcpp.vector cimport vector
+from libc.math cimport fabs, sqrt
cimport numpy as np
@@ -20,37 +18,107 @@ DTYPE = np.double
ctypedef np.double_t DTYPE_t
-@cython.boundscheck(False)
-@cython.wraparound(False)
-@cython.cdivision(True)
-cdef inline double _distance_1d(double[:] x, double[:] y, double[:] z,
- int i, int j) nogil:
+cdef inline double _distance_1d(
+ const double[:] x,
+ const double[:] y,
+ const double[:] z,
+ const int i,
+ const int j
+) nogil:
return sqrt((x[i] - x[j]) * (x[i] - x[j]))
-@cython.boundscheck(False)
-@cython.wraparound(False)
-@cython.cdivision(True)
-cdef inline double _distance_2d(double[:] x, double[:] y, double[:] z,
- int i, int j) nogil:
+cdef inline double _distance_2d(
+ const double[:] x,
+ const double[:] y,
+ const double[:] z,
+ const int i,
+ const int j
+) nogil:
return sqrt((x[i] - x[j]) * (x[i] - x[j]) + (y[i] - y[j]) * (y[i] - y[j]))
-@cython.boundscheck(False)
-@cython.wraparound(False)
-@cython.cdivision(True)
-cdef inline double _distance_3d(double[:] x, double[:] y, double[:] z,
- int i, int j) nogil:
+cdef inline double _distance_3d(
+ const double[:] x,
+ const double[:] y,
+ const double[:] z,
+ const int i,
+ const int j
+) nogil:
return sqrt((x[i] - x[j]) * (x[i] - x[j]) +
(y[i] - y[j]) * (y[i] - y[j]) +
(z[i] - z[j]) * (z[i] - z[j]))
-ctypedef double (*_dist_func)(double[:], double[:], double[:], int, int) nogil
+cdef inline double estimator_matheron(const double f_diff) nogil:
+ return f_diff * f_diff
-@cython.boundscheck(False)
-@cython.wraparound(False)
-@cython.cdivision(True)
-def unstructured(double[:] f, double[:] bin_edges, double[:] x,
- double[:] y=None, double[:] z=None):
+cdef inline double estimator_cressie(const double f_diff) nogil:
+ return sqrt(fabs(f_diff))
+
+ctypedef double (*_estimator_func)(const double) nogil
+
+cdef inline void normalization_matheron(
+ vector[double]& variogram,
+ vector[long]& counts
+):
+ cdef int i
+ for i in range(variogram.size()):
+ # avoid division by zero
+ if counts[i] == 0:
+ counts[i] = 1
+ variogram[i] /= (2. * counts[i])
+
+cdef inline void normalization_cressie(
+ vector[double]& variogram,
+ vector[long]& counts
+):
+ cdef int i
+ for i in range(variogram.size()):
+ # avoid division by zero
+ if counts[i] == 0:
+ counts[i] = 1
+ variogram[i] = (
+ (1./counts[i] * variogram[i])**4 /
+ (0.457 + 0.494 / counts[i] + 0.045 / counts[i]**2)
+ )
+
+ctypedef void (*_normalization_func)(
+ vector[double]&,
+ vector[long]&
+)
+
+cdef _estimator_func choose_estimator_func(str estimator_type):
+ cdef _estimator_func estimator_func
+ if estimator_type == 'm':
+ estimator_func = estimator_matheron
+ elif estimator_type == 'c':
+ estimator_func = estimator_cressie
+ return estimator_func
+
+cdef _normalization_func choose_estimator_normalization(str estimator_type):
+ cdef _normalization_func normalization_func
+ if estimator_type == 'm':
+ normalization_func = normalization_matheron
+ elif estimator_type == 'c':
+ normalization_func = normalization_cressie
+ return normalization_func
+
+ctypedef double (*_dist_func)(
+ const double[:],
+ const double[:],
+ const double[:],
+ const int,
+ const int
+) nogil
+
+
+def unstructured(
+ const double[:] f,
+ const double[:] bin_edges,
+ const double[:] x,
+ const double[:] y=None,
+ const double[:] z=None,
+ str estimator_type='m'
+):
if x.shape[0] != f.shape[0]:
raise ValueError('len(x) = {0} != len(f) = {1} '.
format(x.shape[0], f.shape[0]))
@@ -58,29 +126,34 @@ def unstructured(double[:] f, double[:] bin_edges, double[:] x,
raise ValueError('len(bin_edges) too small')
cdef _dist_func distance
- #3d
+ # 3d
if z is not None:
if z.shape[0] != f.shape[0]:
raise ValueError('len(z) = {0} != len(f) = {1} '.
format(z.shape[0], f.shape[0]))
distance = _distance_3d
- #2d
+ # 2d
elif y is not None:
if y.shape[0] != f.shape[0]:
raise ValueError('len(y) = {0} != len(f) = {1} '.
format(y.shape[0], f.shape[0]))
distance = _distance_2d
- #1d
+ # 1d
else:
distance = _distance_1d
+ cdef _estimator_func estimator_func = choose_estimator_func(estimator_type)
+ cdef _normalization_func normalization_func = (
+ choose_estimator_normalization(estimator_type)
+ )
+
cdef int i_max = bin_edges.shape[0] - 1
cdef int j_max = x.shape[0] - 1
cdef int k_max = x.shape[0]
- cdef double[:] variogram = np.zeros(len(bin_edges)-1)
- cdef long[:] counts = np.zeros_like(variogram, dtype=np.int)
- cdef int i, j, k, d
+ cdef vector[double] variogram = vector[double](len(bin_edges)-1, 0.0)
+ cdef vector[long] counts = vector[long](len(bin_edges)-1, 0)
+ cdef int i, j, k
cdef DTYPE_t dist
for i in prange(i_max, nogil=True):
for j in range(j_max):
@@ -88,158 +161,65 @@ def unstructured(double[:] f, double[:] bin_edges, double[:] x,
dist = distance(x, y, z, k, j)
if dist >= bin_edges[i] and dist < bin_edges[i+1]:
counts[i] += 1
- variogram[i] += (f[k] - f[j])**2
- #avoid division by zero
- for i in range(i_max):
- if counts[i] == 0:
- counts[i] = 1
- variogram[i] /= (2. * counts[i])
+ variogram[i] += estimator_func(f[k] - f[j])
+
+ normalization_func(variogram, counts)
return np.asarray(variogram)
-@cython.boundscheck(False)
-@cython.wraparound(False)
-@cython.cdivision(True)
-def structured_3d(double[:,:,:] f):
+def structured(const double[:,:,:] f, str estimator_type='m'):
+ cdef _estimator_func estimator_func = choose_estimator_func(estimator_type)
+ cdef _normalization_func normalization_func = (
+ choose_estimator_normalization(estimator_type)
+ )
+
cdef int i_max = f.shape[0] - 1
cdef int j_max = f.shape[1]
cdef int k_max = f.shape[2]
cdef int l_max = i_max + 1
- cdef double[:] variogram = np.zeros(l_max)
- cdef long[:] counts = np.zeros_like(variogram, dtype=np.int)
+ cdef vector[double] variogram = vector[double](l_max, 0.0)
+ cdef vector[long] counts = vector[long](l_max, 0)
cdef int i, j, k, l
- for i in prange(i_max, nogil=True):
- for j in range(j_max):
- for k in range(k_max):
- for l in range(1, l_max-i):
- counts[l] += 1
- variogram[l] += (f[i,j,k] - f[i+l,j,k])**2
- #avoid division by zero
- for i in range(l_max):
- if counts[i] == 0:
- counts[i] = 1
- variogram[i] /= (2. * counts[i])
- return np.asarray(variogram)
-
-@cython.boundscheck(False)
-@cython.wraparound(False)
-@cython.cdivision(True)
-def structured_2d(double[:,:] f):
- cdef int i_max = f.shape[0] - 1
- cdef int j_max = f.shape[1]
- cdef int k_max = i_max + 1
-
- cdef double[:] variogram = np.zeros(k_max)
- cdef long[:] counts = np.zeros_like(variogram, dtype=np.int)
- cdef int i, j, k
+ with nogil, parallel():
+ for i in range(i_max):
+ for j in range(j_max):
+ for k in range(k_max):
+ for l in prange(1, l_max-i):
+ counts[l] += 1
+ variogram[l] += estimator_func(f[i,j,k] - f[i+l,j,k])
- for i in prange(i_max, nogil=True):
- for j in range(j_max):
- for k in range(1, k_max-i):
- counts[k] += 1
- variogram[k] += (f[i,j] - f[i+k,j])**2
- #avoid division by zero
- for i in range(k_max):
- if counts[i] == 0:
- counts[i] = 1
- variogram[i] /= (2. * counts[i])
+ normalization_func(variogram, counts)
return np.asarray(variogram)
-@cython.boundscheck(False)
-@cython.wraparound(False)
-@cython.cdivision(True)
-def structured_1d(double[:] f):
- cdef int i_max = f.shape[0] - 1
- cdef int j_max = i_max + 1
-
- cdef double[:] variogram = np.zeros(j_max)
- cdef long[:] counts = np.zeros_like(variogram, dtype=np.int)
- cdef int i, j
-
- for i in range(i_max):
- for j in range(1, j_max-i):
- counts[j] += 1
- variogram[j] += (f[i] - f[i+j])**2
- #avoid division by zero
- for i in range(j_max):
- if counts[i] == 0:
- counts[i] = 1
- variogram[i] /= (2. * counts[i])
- return np.asarray(variogram)
+def ma_structured(
+ const double[:,:,:] f,
+ const bint[:,:,:] mask,
+ str estimator_type='m'
+):
+ cdef _estimator_func estimator_func = choose_estimator_func(estimator_type)
+ cdef _normalization_func normalization_func = (
+ choose_estimator_normalization(estimator_type)
+ )
-@cython.boundscheck(False)
-@cython.wraparound(False)
-@cython.cdivision(True)
-def ma_structured_3d(double[:,:,:] f, bint[:,:,:] mask):
cdef int i_max = f.shape[0] - 1
cdef int j_max = f.shape[1]
cdef int k_max = f.shape[2]
cdef int l_max = i_max + 1
- cdef double[:] variogram = np.zeros(l_max)
- cdef long[:] counts = np.zeros_like(variogram, dtype=np.int)
+ cdef vector[double] variogram = vector[double](l_max, 0.0)
+ cdef vector[long] counts = vector[long](l_max, 0)
cdef int i, j, k, l
- for i in prange(i_max, nogil=True):
- for j in range(j_max):
- for k in range(k_max):
- for l in range(1, l_max-i):
- if not mask[i,j,k] and not mask[i+l,j,k]:
- counts[l] += 1
- variogram[l] += (f[i,j,k] - f[i+l,j,k])**2
- #avoid division by zero
- for i in range(l_max):
- if counts[i] == 0:
- counts[i] = 1
- variogram[i] /= (2. * counts[i])
- return np.asarray(variogram)
-
-@cython.boundscheck(False)
-@cython.wraparound(False)
-@cython.cdivision(True)
-def ma_structured_2d(double[:,:] f, bint[:,:] mask):
- cdef int i_max = f.shape[0] - 1
- cdef int j_max = f.shape[1]
- cdef int k_max = i_max + 1
-
- cdef double[:] variogram = np.zeros(k_max)
- cdef long[:] counts = np.zeros_like(variogram, dtype=np.int)
- cdef int i, j, k
+ with nogil, parallel():
+ for i in range(i_max):
+ for j in range(j_max):
+ for k in range(k_max):
+ for l in prange(1, l_max-i):
+ if not mask[i,j,k] and not mask[i+l,j,k]:
+ counts[l] += 1
+ variogram[l] += estimator_func(f[i,j,k] - f[i+l,j,k])
- for i in prange(i_max, nogil=True):
- for j in range(j_max):
- for k in range(1, k_max-i):
- if not mask[i,j] and not mask[i+k,j]:
- counts[k] += 1
- variogram[k] += (f[i,j] - f[i+k,j])**2
- #avoid division by zero
- for i in range(k_max):
- if counts[i] == 0:
- counts[i] = 1
- variogram[i] /= (2. * counts[i])
- return np.asarray(variogram)
-
-@cython.boundscheck(False)
-@cython.wraparound(False)
-@cython.cdivision(True)
-def ma_structured_1d(double[:] f, bint[:] mask):
- cdef int i_max = f.shape[0] - 1
- cdef int j_max = i_max + 1
-
- cdef double[:] variogram = np.zeros(j_max)
- cdef long[:] counts = np.zeros_like(variogram, dtype=np.int)
- cdef int i, j
-
- for i in range(i_max):
- for j in range(1, j_max-i):
- if not mask[i] and not mask[j]:
- counts[j] += 1
- variogram[j] += (f[i] - f[i+j])**2
- #avoid division by zero
- for i in range(j_max):
- if counts[i] == 0:
- counts[i] = 1
- variogram[i] /= (2. * counts[i])
+ normalization_func(variogram, counts)
return np.asarray(variogram)
diff --git a/gstools/variogram/variogram.py b/gstools/variogram/variogram.py
index b2800fed..ab87562f 100644
--- a/gstools/variogram/variogram.py
+++ b/gstools/variogram/variogram.py
@@ -11,26 +11,35 @@
vario_estimate_structured
"""
# pylint: disable=C0103
-from __future__ import division, absolute_import, print_function
import numpy as np
from gstools.tools.geometric import pos2xyz
-from gstools.variogram.estimator import (
- unstructured,
- structured_3d,
- structured_2d,
- structured_1d,
- ma_structured_3d,
- ma_structured_2d,
- ma_structured_1d,
-)
+from gstools.variogram.estimator import unstructured, structured, ma_structured
__all__ = ["vario_estimate_unstructured", "vario_estimate_structured"]
+def _set_estimator(estimator):
+ """Translate the verbose Python estimator identifier to single char"""
+ if estimator.lower() == "matheron":
+ cython_estimator = "m"
+ elif estimator.lower() == "cressie":
+ cython_estimator = "c"
+ else:
+ raise ValueError(
+ "Unknown variogram estimator function " + str(estimator)
+ )
+ return cython_estimator
+
+
def vario_estimate_unstructured(
- pos, field, bin_edges, sampling_size=None, sampling_seed=None
+ pos,
+ field,
+ bin_edges,
+ sampling_size=None,
+ sampling_seed=None,
+ estimator="matheron",
):
r"""
Estimates the variogram on a unstructured grid.
@@ -38,10 +47,20 @@ def vario_estimate_unstructured(
The algorithm calculates following equation:
.. math::
- \gamma(r_k) = \frac{1}{2 N} \sum_{i=1}^N (z(\mathbf x_i) -
- z(\mathbf x_i'))^2, \; \mathrm{ with}
+ \gamma(r_k) = \frac{1}{2 N(r_k)} \sum_{i=1}^{N(r_k)} (z(\mathbf x_i) -
+ z(\mathbf x_i'))^2 \; ,
+
+ with :math:`r_k \leq \| \mathbf x_i - \mathbf x_i' \| < r_{k+1}` being the bins.
+
+ Or if the estimator "cressie" was chosen:
+
+ .. math::
+ \gamma(r_k) = \frac{\left(\frac{1}{N(r_k)} \sum_{i=1}^{N(r_k)}
+ \left|z(\mathbf x_i) - z(\mathbf x_i')\right|^{0.5}\right)^4}
+ {0.457 + 0.494 / N(r_k) + 0.045 / N^2(r_k)} \; ,
- r_k \leq \| \mathbf x_i - \mathbf x_i' \| < r_{k+1}
+ with :math:`r_k \leq \| \mathbf x_i - \mathbf x_i' \| < r_{k+1}` being the bins.
+ The Cressie estimator is more robust to outliers.
Notes
-----
@@ -64,6 +83,13 @@ def vario_estimate_unstructured(
sampling_seed : :class:`int` or :any:`None`, optional
seed for samples if sampling_size is given.
Default: :any:`None`
+ estimator : :class:`str`, optional
+ the estimator function, possible choices:
+
+ * "matheron": the standard method of moments of Matheron
+ * "cressie": an estimator more robust to outliers
+
+ Default: "matheron"
Returns
-------
@@ -87,20 +113,37 @@ def vario_estimate_unstructured(
if dim > 2:
z = z[sampled_idx]
- return bin_centres, unstructured(field, bin_edges, x, y, z)
+ cython_estimator = _set_estimator(estimator)
+ return (
+ bin_centres,
+ unstructured(
+ field, bin_edges, x, y, z, estimator_type=cython_estimator
+ ),
+ )
-def vario_estimate_structured(field, direction="x"):
+
+def vario_estimate_structured(field, direction="x", estimator="matheron"):
r"""Estimates the variogram on a regular grid.
The indices of the given direction are used for the bins.
The algorithm calculates following equation:
.. math::
- \gamma(r_k) = \frac{1}{2 N} \sum_{i=1}^N (z(\mathbf x_i) -
- z(\mathbf x_i'))^2, \; \mathrm{ with}
+ \gamma(r_k) = \frac{1}{2 N(r_k)} \sum_{i=1}^{N(r_k)} (z(\mathbf x_i) -
+ z(\mathbf x_i'))^2 \; ,
+
+ with :math:`r_k \leq \| \mathbf x_i - \mathbf x_i' \| < r_{k+1}` being the bins.
+
+ Or if the estimator "cressie" was chosen:
+
+ .. math::
+ \gamma(r_k) = \frac{\left(\frac{1}{N(r_k)} \sum_{i=1}^{N(r_k)}
+ \left|z(\mathbf x_i) - z(\mathbf x_i')\right|^{0.5}\right)^4}
+ {0.457 + 0.494 / N(r_k) + 0.045 / N^2(r_k)} \; ,
- r_k \leq \| \mathbf x_i - \mathbf x_i' \| < r_{k+1}
+ with :math:`r_k \leq \| \mathbf x_i - \mathbf x_i' \| < r_{k+1}` being the bins.
+ The Cressie estimator is more robust to outliers.
Warnings
--------
@@ -116,6 +159,13 @@ def vario_estimate_structured(field, direction="x"):
the spatially distributed data
direction : :class:`str`
the axis over which the variogram will be estimated (x, y, z)
+ estimator : :class:`str`, optional
+ the estimator function, possible choices:
+
+ * "mathoron": the standard method of moments of Matheron
+ * "cressie": an estimator more robust to outliers
+
+ Default: "matheron"
Returns
-------
@@ -130,7 +180,6 @@ def vario_estimate_structured(field, direction="x"):
mask = None
field = np.array(field, ndmin=1, dtype=np.double)
masked = False
- shape = field.shape
if direction == "x":
axis_to_swap = 0
@@ -145,19 +194,17 @@ def vario_estimate_structured(field, direction="x"):
if masked:
mask = mask.swapaxes(0, axis_to_swap)
- if len(shape) == 3:
- if mask is None:
- gamma = structured_3d(field)
- else:
- gamma = ma_structured_3d(field, mask)
- elif len(shape) == 2:
- if mask is None:
- gamma = structured_2d(field)
- else:
- gamma = ma_structured_2d(field, mask)
+ cython_estimator = _set_estimator(estimator)
+
+ # fill up the field with empty dimensions up to a number of 3
+ for i in range(3 - len(field.shape)):
+ field = field[..., np.newaxis]
+ if masked:
+ for i in range(3 - len(mask.shape)):
+ mask = mask[..., np.newaxis]
+
+ if mask is None:
+ gamma = structured(field, cython_estimator)
else:
- if mask is None:
- gamma = structured_1d(np.array(field, ndmin=1, dtype=np.double))
- else:
- gamma = ma_structured_1d(field, mask)
+ gamma = ma_structured(field, mask, cython_estimator)
return gamma
diff --git a/requirements.txt b/requirements.txt
index da14c557..56c77216 100644
--- a/requirements.txt
+++ b/requirements.txt
@@ -1,6 +1,5 @@
numpy>=1.14.5
scipy>=1.1.0
-hankel>=0.3.6
+hankel>=1.0.2
emcee>=3.0.0
-pyevtk
-six
+pyevtk>=1.1.1
diff --git a/requirements_setup.txt b/requirements_setup.txt
new file mode 100755
index 00000000..7329b9c1
--- /dev/null
+++ b/requirements_setup.txt
@@ -0,0 +1,4 @@
+setuptools>=41.0.1
+setuptools_scm>=3.5.0
+cython>=0.28.3
+numpy>=1.14.5
\ No newline at end of file
diff --git a/requirements_test.txt b/requirements_test.txt
new file mode 100755
index 00000000..be10813e
--- /dev/null
+++ b/requirements_test.txt
@@ -0,0 +1,2 @@
+pytest-cov>=2.8.0
+pytest>=5.3.0
diff --git a/setup.cfg b/setup.cfg
index c62da7f2..f48fdadb 100644
--- a/setup.cfg
+++ b/setup.cfg
@@ -1,6 +1,3 @@
[metadata]
description-file = README.md
license_file = LICENSE
-
-[bdist_wheel]
-universal = 1
diff --git a/setup.py b/setup.py
index e755f698..eadc302c 100644
--- a/setup.py
+++ b/setup.py
@@ -1,7 +1,10 @@
# -*- coding: utf-8 -*-
"""GSTools: A geostatistical toolbox."""
-from __future__ import division, absolute_import, print_function
-import sys, os, codecs, re, tempfile, glob, subprocess, shutil
+import sys
+import os
+import glob
+import tempfile
+import subprocess
from distutils.errors import CompileError, LinkError
from distutils.ccompiler import new_compiler
@@ -9,52 +12,18 @@
from setuptools import setup, find_packages, Distribution, Extension
from Cython.Build import cythonize
-import numpy
+import numpy as np
HERE = os.path.abspath(os.path.dirname(__file__))
-# version finder ##############################################################
-
-
-def read(*file_paths):
- """Read file data."""
- with codecs.open(os.path.join(HERE, *file_paths), "r") as file_in:
- return file_in.read()
-
-
-def find_version(*file_paths):
- """Find version without importing module."""
- version_file = read(*file_paths)
- version_match = re.search(
- r"^__version__ = ['\"]([^'\"]*)['\"]", version_file, re.M
- )
- if version_match:
- return version_match.group(1)
- raise RuntimeError("Unable to find version string.")
-
-
# openmp finder ###############################################################
# This code is adapted for a large part from the scikit-learn openmp helpers,
# which can be found at:
# https://github.com/scikit-learn/scikit-learn/blob/master/sklearn/_build_utils/openmp_helpers.py
-# TemporaryDirectory not avialable in python2
-class _TemporaryDirectory(object):
- def __enter__(self):
- self.dir_name = tempfile.mkdtemp()
- return self.dir_name
-
- def __exit__(self, exc_type, exc_value, traceback):
- shutil.rmtree(self.dir_name)
-
-
-TemporaryDirectory = getattr(
- tempfile, "TemporaryDirectory", _TemporaryDirectory
-)
-
CCODE = """
#include
#include
@@ -101,7 +70,7 @@ def check_openmp_support():
ccompiler = new_compiler()
customize_compiler(ccompiler)
- with TemporaryDirectory() as tmp_dir:
+ with tempfile.TemporaryDirectory() as tmp_dir:
try:
os.chdir(tmp_dir)
# Write test program
@@ -186,7 +155,7 @@ class MPDistribution(Distribution):
Extension(
"gstools.field.summator",
[os.path.join("gstools", "field", "summator.pyx")],
- include_dirs=[numpy.get_include()],
+ include_dirs=[np.get_include()],
extra_compile_args=FLAGS,
extra_link_args=FLAGS,
)
@@ -195,7 +164,8 @@ class MPDistribution(Distribution):
Extension(
"gstools.variogram.estimator",
[os.path.join("gstools", "variogram", "estimator.pyx")],
- include_dirs=[numpy.get_include()],
+ language="c++",
+ include_dirs=[np.get_include()],
extra_compile_args=FLAGS,
extra_link_args=FLAGS,
)
@@ -204,7 +174,7 @@ class MPDistribution(Distribution):
Extension(
"gstools.krige.krigesum",
[os.path.join("gstools", "krige", "krigesum.pyx")],
- include_dirs=[numpy.get_include()],
+ include_dirs=[np.get_include()],
extra_compile_args=FLAGS,
extra_link_args=FLAGS,
)
@@ -218,16 +188,24 @@ class MPDistribution(Distribution):
for ext_m in EXT_MODULES:
ext_m.cython_directives = {"embedsignature": True}
-
# setup #######################################################################
+with open(os.path.join(HERE, "README.md"), encoding="utf-8") as f:
+ README = f.read()
+with open(os.path.join(HERE, "requirements.txt"), encoding="utf-8") as f:
+ REQ = f.read().splitlines()
+with open(os.path.join(HERE, "requirements_setup.txt"), encoding="utf-8") as f:
+ REQ_SETUP = f.read().splitlines()
+with open(os.path.join(HERE, "requirements_test.txt"), encoding="utf-8") as f:
+ REQ_TEST = f.read().splitlines()
+with open(
+ os.path.join(HERE, "docs", "requirements_doc.txt"), encoding="utf-8"
+) as f:
+ REQ_DOC = f.read().splitlines()
+
+REQ_DEV = REQ_SETUP + REQ_TEST + REQ_DOC
-# version import not possible due to cython
-# see: https://packaging.python.org/guides/single-sourcing-package-version/
-VERSION = find_version("gstools", "_version.py")
DOCLINE = __doc__.split("\n")[0]
-README = read("README.md")
-
CLASSIFIERS = [
"Development Status :: 5 - Production/Stable",
"Intended Audience :: Developers",
@@ -237,20 +215,23 @@ class MPDistribution(Distribution):
"Natural Language :: English",
"Operating System :: Unix",
"Programming Language :: Python",
- "Programming Language :: Python :: 2",
"Programming Language :: Python :: 3",
+ "Programming Language :: Python :: 3.5",
+ "Programming Language :: Python :: 3.6",
+ "Programming Language :: Python :: 3.7",
+ "Programming Language :: Python :: 3.8",
+ "Programming Language :: Python :: 3 :: Only",
"Topic :: Scientific/Engineering",
"Topic :: Utilities",
]
setup(
name="gstools",
- version=VERSION,
- maintainer="Lennart Schueler, Sebastian Mueller",
- maintainer_email="info@geostat-framework.org",
description=DOCLINE,
long_description=README,
long_description_content_type="text/markdown",
+ maintainer="Lennart Schueler, Sebastian Mueller",
+ maintainer_email="info@geostat-framework.org",
author="Lennart Schueler, Sebastian Mueller",
author_email="info@geostat-framework.org",
url="https://github.com/GeoStat-Framework/GSTools",
@@ -258,18 +239,24 @@ class MPDistribution(Distribution):
classifiers=CLASSIFIERS,
platforms=["Windows", "Linux", "Solaris", "Mac OS-X", "Unix"],
include_package_data=True,
- setup_requires=["numpy>=1.14.5", "cython>=0.28.3", "setuptools>=41.0.1"],
- install_requires=[
- "numpy>=1.14.5",
- "scipy>=1.1.0",
- "hankel>=0.3.6",
- "emcee>=3.0.0",
- "pyevtk",
- "six",
- ],
- extras_require={"plotting": ["pyvista", "matplotlib"]},
+ python_requires=">=3.5",
+ use_scm_version={
+ "relative_to": __file__,
+ "write_to": "gstools/_version.py",
+ "write_to_template": "__version__ = '{version}'",
+ "local_scheme": "no-local-version",
+ "fallback_version": "0.0.0.dev0",
+ },
+ setup_requires=REQ_SETUP,
+ install_requires=REQ,
+ extras_require={
+ "plotting": ["pyvista", "matplotlib"],
+ "doc": REQ_DOC,
+ "test": REQ_TEST,
+ "dev": REQ_DEV,
+ },
packages=find_packages(exclude=["tests*", "docs*"]),
ext_modules=EXT_MODULES,
- include_dirs=[numpy.get_include()],
+ include_dirs=[np.get_include()],
distclass=MPDistribution,
)
diff --git a/tests/__init__.py b/tests/__init__.py
deleted file mode 100644
index e69de29b..00000000
diff --git a/tests/test_condition.py b/tests/test_condition.py
index e04a89cb..e7c58879 100644
--- a/tests/test_condition.py
+++ b/tests/test_condition.py
@@ -2,7 +2,6 @@
"""
This is the unittest of CovModel class.
"""
-from __future__ import division, absolute_import, print_function
import numpy as np
import unittest
diff --git a/tests/test_covmodel.py b/tests/test_covmodel.py
index 9c8345ce..d331773d 100644
--- a/tests/test_covmodel.py
+++ b/tests/test_covmodel.py
@@ -2,8 +2,7 @@
"""
This is the unittest of CovModel class.
"""
-from __future__ import division, absolute_import, print_function
-
+import numpy as np
import unittest
from gstools import (
CovModel,
@@ -60,6 +59,13 @@ def test_creation(self):
with self.assertRaises(TypeError):
CovModel()
+ class User(CovModel):
+ def cor(self, h):
+ return np.exp(-h ** 2)
+
+ user = User(len_scale=2)
+ self.assertAlmostEqual(user.correlation(1), np.exp(-0.25))
+
for Model in self.cov_models:
for dim in self.dims:
for angles in self.angles:
diff --git a/tests/test_incomprrandmeth.py b/tests/test_incomprrandmeth.py
index ea31bd2e..2ef39d5c 100644
--- a/tests/test_incomprrandmeth.py
+++ b/tests/test_incomprrandmeth.py
@@ -2,7 +2,6 @@
"""
This is the unittest of the RandMeth class.
"""
-from __future__ import division, absolute_import, print_function
import copy
import unittest
@@ -13,9 +12,7 @@
class TestIncomprRandMeth(unittest.TestCase):
def setUp(self):
- self.cov_model_2d = Gaussian(
- dim=2, var=1.5, len_scale=2.5, mode_no=100
- )
+ self.cov_model_2d = Gaussian(dim=2, var=1.5, len_scale=2.5)
self.cov_model_3d = copy.deepcopy(self.cov_model_2d)
self.cov_model_3d.dim = 3
self.seed = 19031977
@@ -77,7 +74,7 @@ def test_struct_unstruct(self):
k += 1
def test_assertions(self):
- cov_model_1d = Gaussian(dim=1, var=1.5, len_scale=2.5, mode_no=100)
+ cov_model_1d = Gaussian(dim=1, var=1.5, len_scale=2.5)
self.assertRaises(ValueError, IncomprRandMeth, cov_model_1d)
diff --git a/tests/test_krige.py b/tests/test_krige.py
index 370b7b02..3afee2b0 100644
--- a/tests/test_krige.py
+++ b/tests/test_krige.py
@@ -1,12 +1,15 @@
# -*- coding: utf-8 -*-
"""
-This is the unittest of CovModel class.
+This is the unittest of the kriging module.
"""
-from __future__ import division, absolute_import, print_function
import numpy as np
import unittest
-from gstools import Gaussian, Exponential, Spherical, krige
+from gstools import Gaussian, Exponential, Spherical, krige, SRF
+
+
+def trend(*xyz):
+ return xyz[0]
class TestKrige(unittest.TestCase):
@@ -22,79 +25,173 @@ def setUp(self):
[4.7, 3.8, 2.5, 1.74],
]
)
+ # indices for the date in the grid
+ self.data_idx = tuple(np.array(self.data[:, :3] * 10, dtype=int).T)
+ # x, y, z componentes for the conditon position
self.cond_pos = (self.data[:, 0], self.data[:, 1], self.data[:, 2])
+ # condition values
self.cond_val = self.data[:, 3]
+ # the arithmetic mean of the conditions
self.mean = np.mean(self.cond_val)
- self.grid_x = np.concatenate((self.cond_pos[0], np.linspace(5, 20)))
- self.grid_y = np.concatenate((self.cond_pos[1], np.linspace(5, 20)))
- self.grid_z = np.concatenate((self.cond_pos[2], np.linspace(5, 20)))
- self.pos = (self.grid_x, self.grid_y, self.grid_z)
+ # the grid
+ self.x = np.linspace(0, 5, 51)
+ self.y = np.linspace(0, 6, 61)
+ self.z = np.linspace(0, 7, 71)
+ self.pos = (self.x, self.y, self.z)
+ self.grids = [self.x]
+ self.grids.append(np.meshgrid(self.x, self.y, indexing="ij"))
+ self.grids.append(np.meshgrid(self.x, self.y, self.z, indexing="ij"))
+ self.grid_shape = [51, 61, 71]
def test_simple(self):
for Model in self.cov_models:
- model = Model(
- dim=1, var=0.5, len_scale=2, anis=[0.1, 1], angles=[0.5, 0, 0]
- )
- simple = krige.Simple(
- model, self.mean, self.cond_pos[0], self.cond_val
- )
- field_1, __ = simple.unstructured(self.pos[0])
- field_2, __ = simple.structured(self.pos[0])
- for i, val in enumerate(self.cond_val):
- self.assertAlmostEqual(val, field_1[i], places=2)
- self.assertAlmostEqual(val, field_2[(i,)], places=2)
- self.assertAlmostEqual(self.mean, field_1[-1], places=2)
- self.assertAlmostEqual(self.mean, field_2[(-1,)], places=2)
-
- for dim in self.dims[1:]:
+ for dim in self.dims:
model = Model(
dim=dim,
- var=0.5,
+ var=2,
len_scale=2,
- anis=[0.1, 1],
- angles=[0.5, 0, 0],
+ anis=[0.9, 0.8],
+ angles=[2, 1, 0.5],
)
simple = krige.Simple(
- model, self.mean, self.cond_pos[:dim], self.cond_val
+ model, self.cond_pos[:dim], self.cond_val, self.mean
)
- field_1, __ = simple.unstructured(self.pos[:dim])
+ field_1, __ = simple.unstructured(self.grids[dim - 1])
+ field_1 = field_1.reshape(self.grid_shape[:dim])
field_2, __ = simple.structured(self.pos[:dim])
- for i, val in enumerate(self.cond_val):
- self.assertAlmostEqual(val, field_1[i], places=2)
- self.assertAlmostEqual(val, field_2[dim * (i,)], places=2)
- self.assertAlmostEqual(self.mean, field_1[-1], places=2)
self.assertAlmostEqual(
- self.mean, field_2[dim * (-1,)], places=2
+ np.max(np.abs(field_1 - field_2)), 0.0, places=2
)
+ for i, val in enumerate(self.cond_val):
+ self.assertAlmostEqual(
+ field_1[self.data_idx[:dim]][i], val, places=2
+ )
def test_ordinary(self):
+ for trend_func in [None, trend]:
+ for Model in self.cov_models:
+ for dim in self.dims:
+ model = Model(
+ dim=dim,
+ var=5,
+ len_scale=10,
+ anis=[0.9, 0.8],
+ angles=[2, 1, 0.5],
+ )
+ ordinary = krige.Ordinary(
+ model, self.cond_pos[:dim], self.cond_val, trend_func
+ )
+ field_1, __ = ordinary.unstructured(self.grids[dim - 1])
+ field_1 = field_1.reshape(self.grid_shape[:dim])
+ field_2, __ = ordinary.structured(self.pos[:dim])
+ self.assertAlmostEqual(
+ np.max(np.abs(field_1 - field_2)), 0.0, places=2
+ )
+ for i, val in enumerate(self.cond_val):
+ self.assertAlmostEqual(
+ field_1[self.data_idx[:dim]][i], val, places=2
+ )
+
+ def test_universal(self):
+ # "quad" -> to few conditional points
+ for drift in ["linear", 0, 1, trend]:
+ for Model in self.cov_models:
+ for dim in self.dims:
+ model = Model(
+ dim=dim,
+ var=2,
+ len_scale=10,
+ anis=[0.9, 0.8],
+ angles=[2, 1, 0.5],
+ )
+ universal = krige.Universal(
+ model, self.cond_pos[:dim], self.cond_val, drift
+ )
+ field_1, __ = universal.unstructured(self.grids[dim - 1])
+ field_1 = field_1.reshape(self.grid_shape[:dim])
+ field_2, __ = universal.structured(self.pos[:dim])
+ self.assertAlmostEqual(
+ np.max(np.abs(field_1 - field_2)), 0.0, places=2
+ )
+ for i, val in enumerate(self.cond_val):
+ self.assertAlmostEqual(
+ field_2[self.data_idx[:dim]][i], val, places=2
+ )
+
+ def test_detrended(self):
+
for Model in self.cov_models:
- model = Model(
- dim=1, var=0.5, len_scale=2, anis=[0.1, 1], angles=[0.5, 0, 0]
+ for dim in self.dims:
+ model = Model(
+ dim=dim,
+ var=2,
+ len_scale=10,
+ anis=[0.5, 0.2],
+ angles=[0.4, 0.2, 0.1],
+ )
+ detrended = krige.Detrended(
+ model, self.cond_pos[:dim], self.cond_val, trend
+ )
+ field_1, __ = detrended.unstructured(self.grids[dim - 1])
+ field_1 = field_1.reshape(self.grid_shape[:dim])
+ field_2, __ = detrended.structured(self.pos[:dim])
+ # detrended.plot()
+ self.assertAlmostEqual(
+ np.max(np.abs(field_1 - field_2)), 0.0, places=2
+ )
+ for i, val in enumerate(self.cond_val):
+ self.assertAlmostEqual(
+ field_2[self.data_idx[:dim]][i], val, places=2
+ )
+
+ def test_extdrift(self):
+ ext_drift = []
+ cond_drift = []
+ for i, grid in enumerate(self.grids):
+ dim = i + 1
+ model = Exponential(
+ dim=dim,
+ var=2,
+ len_scale=10,
+ anis=[0.9, 0.8],
+ angles=[2, 1, 0.5],
)
- ordinary = krige.Ordinary(model, self.cond_pos[0], self.cond_val)
- field_1, __ = ordinary.unstructured(self.pos[0])
- field_2, __ = ordinary.structured(self.pos[0])
- for i, val in enumerate(self.cond_val):
- self.assertAlmostEqual(val, field_1[i], places=2)
- self.assertAlmostEqual(val, field_2[(i,)], places=2)
+ srf = SRF(model)
+ field = srf(grid)
+ ext_drift.append(field)
+ field = field.reshape(self.grid_shape[:dim])
+ cond_drift.append(field[self.data_idx[:dim]])
- for dim in self.dims[1:]:
+ for Model in self.cov_models:
+ for dim in self.dims:
model = Model(
dim=dim,
- var=0.5,
- len_scale=2,
- anis=[0.1, 1],
- angles=[0.5, 0, 0],
+ var=2,
+ len_scale=10,
+ anis=[0.5, 0.2],
+ angles=[0.4, 0.2, 0.1],
+ )
+ extdrift = krige.ExtDrift(
+ model,
+ self.cond_pos[:dim],
+ self.cond_val,
+ cond_drift[dim - 1],
)
- ordinary = krige.Ordinary(
- model, self.cond_pos[:dim], self.cond_val
+ field_1, __ = extdrift.unstructured(
+ self.grids[dim - 1], ext_drift=ext_drift[dim - 1]
+ )
+ field_1 = field_1.reshape(self.grid_shape[:dim])
+ field_2, __ = extdrift.structured(
+ self.pos[:dim], ext_drift=ext_drift[dim - 1]
+ )
+ # extdrift.plot()
+ self.assertAlmostEqual(
+ np.max(np.abs(field_1 - field_2)), 0.0, places=2
)
- field_1, __ = ordinary.unstructured(self.pos[:dim])
- field_2, __ = ordinary.structured(self.pos[:dim])
for i, val in enumerate(self.cond_val):
- self.assertAlmostEqual(val, field_1[i], places=2)
- self.assertAlmostEqual(val, field_2[dim * (i,)], places=2)
+ self.assertAlmostEqual(
+ field_2[self.data_idx[:dim]][i], val, places=2
+ )
if __name__ == "__main__":
diff --git a/tests/test_randmeth.py b/tests/test_randmeth.py
index 4846585a..85b2e691 100644
--- a/tests/test_randmeth.py
+++ b/tests/test_randmeth.py
@@ -2,7 +2,6 @@
"""
This is the unittest of the RandMeth class.
"""
-from __future__ import division, absolute_import, print_function
import copy
import unittest
@@ -13,9 +12,7 @@
class TestRandMeth(unittest.TestCase):
def setUp(self):
- self.cov_model_1d = Gaussian(
- dim=1, var=1.5, len_scale=3.5, mode_no=100
- )
+ self.cov_model_1d = Gaussian(dim=1, var=1.5, len_scale=3.5)
self.cov_model_2d = copy.deepcopy(self.cov_model_1d)
self.cov_model_2d.dim = 2
self.cov_model_3d = copy.deepcopy(self.cov_model_1d)
diff --git a/tests/test_rng.py b/tests/test_rng.py
index 9f4736a0..2c00b1ea 100644
--- a/tests/test_rng.py
+++ b/tests/test_rng.py
@@ -2,7 +2,6 @@
"""
This is the unittest of the RNG class.
"""
-from __future__ import division, absolute_import, print_function
import unittest
import numpy as np
diff --git a/tests/test_srf.py b/tests/test_srf.py
index a362d3e1..082a7ec3 100644
--- a/tests/test_srf.py
+++ b/tests/test_srf.py
@@ -3,7 +3,6 @@
"""
This is the unittest of SRF class.
"""
-from __future__ import division, absolute_import, print_function
import unittest
import numpy as np
@@ -13,7 +12,7 @@
class TestSRF(unittest.TestCase):
def setUp(self):
- self.cov_model = Gaussian(dim=2, var=1.5, len_scale=4.0, mode_no=100)
+ self.cov_model = Gaussian(dim=2, var=1.5, len_scale=4.0)
self.mean = 0.3
self.mode_no = 100
@@ -156,7 +155,7 @@ def test_rotation_struct_2d(self):
def test_rotation_unstruct_3d(self):
self.cov_model = Gaussian(
- dim=3, var=1.5, len_scale=4.0, anis=(0.25, 0.5), mode_no=100
+ dim=3, var=1.5, len_scale=4.0, anis=(0.25, 0.5)
)
x_len = len(self.x_grid_c)
y_len = len(self.y_grid_c)
@@ -251,9 +250,28 @@ def test_transform(self):
tf.binary(srf)
srf((self.x_grid, self.y_grid), seed=self.seed, mesh_type="structured")
tf.boxcox(srf)
+ srf((self.x_grid, self.y_grid), seed=self.seed, mesh_type="structured")
+ values = np.linspace(np.min(srf.field), np.max(srf.field), 3)
+ tf.discrete(srf, values)
+
+ srf((self.x_grid, self.y_grid), seed=self.seed, mesh_type="structured")
+ values = [-1, 0, 1]
+ thresholds = [-0.9, 0.1]
+ tf.discrete(srf, values, thresholds)
+ np.testing.assert_array_equal(np.unique(srf.field), [-1, 0, 1])
+
+ srf((self.x_grid, self.y_grid), seed=self.seed, mesh_type="structured")
+ values = [-1, 0, 1]
+ tf.discrete(srf, values, thresholds="arithmetic")
+ np.testing.assert_array_equal(np.unique(srf.field), [-1.0, 0.0, 1.0])
+
+ srf((self.x_grid, self.y_grid), seed=self.seed, mesh_type="structured")
+ values = [-1, 0, 0.5, 1]
+ tf.discrete(srf, values, thresholds="equal")
+ np.testing.assert_array_equal(np.unique(srf.field), values)
def test_incomprrandmeth(self):
- self.cov_model = Gaussian(dim=2, var=0.5, len_scale=1.0, mode_no=100)
+ self.cov_model = Gaussian(dim=2, var=0.5, len_scale=1.0)
srf = SRF(
self.cov_model,
mean=self.mean,
diff --git a/tests/test_variogram_structured.py b/tests/test_variogram_structured.py
index 05debc6c..390ab5b3 100644
--- a/tests/test_variogram_structured.py
+++ b/tests/test_variogram_structured.py
@@ -2,20 +2,11 @@
"""
This is a unittest of the variogram module.
"""
-from __future__ import division, absolute_import, print_function
-import sys
import unittest
import numpy as np
from gstools import variogram
-PY3 = sys.version_info[0] == 3
-# in python3 "long" was replaced with "int"
-if PY3:
- LONGTYPE = int
-else:
- LONGTYPE = long
-
class TestVariogramstructured(unittest.TestCase):
def setUp(self):
@@ -26,63 +17,41 @@ def test_doubles(self):
(41.2, 40.2, 39.7, 39.2, 40.1, 38.3, 39.1, 40.0, 41.1, 40.3),
dtype=np.double,
)
- try:
- gamma = variogram.vario_estimate_structured(z)
- self.assertAlmostEqual(gamma[1], 0.4917, places=4)
- except NotImplementedError as e:
- pass
+ gamma = variogram.vario_estimate_structured(z)
+ self.assertAlmostEqual(gamma[1], 0.4917, places=4)
def test_ints(self):
z = np.array((10, 20, 30, 40), dtype=int)
- try:
- gamma = variogram.vario_estimate_structured(z)
- self.assertAlmostEqual(gamma[1], 50.0, places=4)
- except NotImplementedError as e:
- pass
-
- def test_longs(self):
- z = np.array((10, 20, 30, 40), dtype=LONGTYPE)
- try:
- gamma = variogram.vario_estimate_structured(z)
- self.assertAlmostEqual(gamma[1], 50.0, places=4)
- except NotImplementedError as e:
- pass
+ gamma = variogram.vario_estimate_structured(z)
+ self.assertAlmostEqual(gamma[1], 50.0, places=4)
def test_np_int(self):
z = np.array((10, 20, 30, 40), dtype=np.int)
- try:
- gamma = variogram.vario_estimate_structured(z)
- self.assertAlmostEqual(gamma[1], 50.0, places=4)
- except NotImplementedError as e:
- pass
+ gamma = variogram.vario_estimate_structured(z)
+ self.assertAlmostEqual(gamma[1], 50.0, places=4)
def test_mixed(self):
z = np.array(
(41.2, 40.2, 39.7, 39.2, 40.1, 38.3, 39.1, 40.0, 41.1, 40.3),
dtype=np.double,
)
- try:
- gamma = variogram.vario_estimate_structured(z)
- self.assertAlmostEqual(gamma[1], 0.4917, places=4)
-
- z = np.array((10, 20, 30, 40), dtype=LONGTYPE)
+ gamma = variogram.vario_estimate_structured(z)
+ self.assertAlmostEqual(gamma[1], 0.4917, places=4)
- gamma = variogram.vario_estimate_structured(z)
- self.assertAlmostEqual(gamma[1], 50.0, places=4)
+ z = np.array((10, 20, 30, 40), dtype=int)
- z = np.array((10, 20, 30, 40), dtype=LONGTYPE)
- gamma = variogram.vario_estimate_structured(z)
- self.assertAlmostEqual(gamma[1], 50.0, places=4)
- except NotImplementedError as e:
- pass
+ gamma = variogram.vario_estimate_structured(z)
+ self.assertAlmostEqual(gamma[1], 50.0, places=4)
def test_list(self):
z = [41.2, 40.2, 39.7, 39.2, 40.1, 38.3, 39.1, 40.0, 41.1, 40.3]
- try:
- gamma = variogram.vario_estimate_structured(z)
- self.assertAlmostEqual(gamma[1], 0.4917, places=4)
- except NotImplementedError as e:
- pass
+ gamma = variogram.vario_estimate_structured(z)
+ self.assertAlmostEqual(gamma[1], 0.4917, places=4)
+
+ def test_cressie_1d(self):
+ z = [41.2, 40.2, 39.7, 39.2, 40.1, 38.3, 39.1, 40.0, 41.1, 40.3]
+ gamma = variogram.vario_estimate_structured(z, estimator="cressie")
+ self.assertAlmostEqual(gamma[1], 1.546, places=3)
def test_1d(self):
# literature values
@@ -90,13 +59,10 @@ def test_1d(self):
(41.2, 40.2, 39.7, 39.2, 40.1, 38.3, 39.1, 40.0, 41.1, 40.3),
dtype=np.double,
)
- try:
- gamma = variogram.vario_estimate_structured(z)
- self.assertAlmostEqual(gamma[0], 0.0000, places=4)
- self.assertAlmostEqual(gamma[1], 0.4917, places=4)
- self.assertAlmostEqual(gamma[2], 0.7625, places=4)
- except NotImplementedError as e:
- pass
+ gamma = variogram.vario_estimate_structured(z)
+ self.assertAlmostEqual(gamma[0], 0.0000, places=4)
+ self.assertAlmostEqual(gamma[1], 0.4917, places=4)
+ self.assertAlmostEqual(gamma[2], 0.7625, places=4)
def test_masked_1d(self):
# literature values
@@ -105,18 +71,15 @@ def test_masked_1d(self):
dtype=np.double,
)
z_ma = np.ma.masked_array(z, mask=[0, 0, 0, 0, 0, 0, 0, 0, 0, 0])
- try:
- gamma = variogram.vario_estimate_structured(z_ma)
- self.assertAlmostEqual(gamma[0], 0.0000, places=4)
- self.assertAlmostEqual(gamma[1], 0.4917, places=4)
- self.assertAlmostEqual(gamma[2], 0.7625, places=4)
- z_ma = np.ma.masked_array(z, mask=[1, 0, 0, 0, 0, 0, 0, 0, 0, 0])
- gamma = variogram.vario_estimate_structured(z_ma)
- self.assertAlmostEqual(gamma[0], 0.0000, places=4)
- self.assertAlmostEqual(gamma[1], 0.4906, places=4)
- self.assertAlmostEqual(gamma[2], 0.7107, places=4)
- except NotImplementedError as e:
- pass
+ gamma = variogram.vario_estimate_structured(z_ma)
+ self.assertAlmostEqual(gamma[0], 0.0000, places=4)
+ self.assertAlmostEqual(gamma[1], 0.4917, places=4)
+ self.assertAlmostEqual(gamma[2], 0.7625, places=4)
+ z_ma = np.ma.masked_array(z, mask=[1, 0, 0, 0, 0, 0, 0, 0, 0, 0])
+ gamma = variogram.vario_estimate_structured(z_ma)
+ self.assertAlmostEqual(gamma[0], 0.0000, places=4)
+ self.assertAlmostEqual(gamma[1], 0.4906, places=4)
+ self.assertAlmostEqual(gamma[2], 0.7107, places=4)
def test_masked_2d(self):
rng = np.random.RandomState(1479373475)
@@ -124,35 +87,24 @@ def test_masked_2d(self):
mask = np.zeros_like(field)
field_ma = np.ma.masked_array(field, mask=mask)
- try:
- gamma_x = variogram.vario_estimate_structured(
- field_ma, direction="x"
- )
- gamma_y = variogram.vario_estimate_structured(
- field_ma, direction="y"
- )
-
- var = 1.0 / 12.0
- self.assertAlmostEqual(gamma_x[0], 0.0, places=2)
- self.assertAlmostEqual(gamma_x[len(gamma_x) // 2], var, places=2)
- self.assertAlmostEqual(gamma_x[-1], var, places=2)
- self.assertAlmostEqual(gamma_y[0], 0.0, places=2)
- self.assertAlmostEqual(gamma_y[len(gamma_y) // 2], var, places=2)
- self.assertAlmostEqual(gamma_y[-1], var, places=2)
-
- mask = np.zeros_like(field)
- mask[0, 0] = 1
- field = np.ma.masked_array(field, mask=mask)
- gamma_x = variogram.vario_estimate_structured(
- field_ma, direction="x"
- )
- gamma_y = variogram.vario_estimate_structured(
- field_ma, direction="y"
- )
- self.assertAlmostEqual(gamma_x[0], 0.0, places=2)
- self.assertAlmostEqual(gamma_y[0], 0.0, places=2)
- except NotImplementedError as e:
- pass
+ gamma_x = variogram.vario_estimate_structured(field_ma, direction="x")
+ gamma_y = variogram.vario_estimate_structured(field_ma, direction="y")
+
+ var = 1.0 / 12.0
+ self.assertAlmostEqual(gamma_x[0], 0.0, places=2)
+ self.assertAlmostEqual(gamma_x[len(gamma_x) // 2], var, places=2)
+ self.assertAlmostEqual(gamma_x[-1], var, places=2)
+ self.assertAlmostEqual(gamma_y[0], 0.0, places=2)
+ self.assertAlmostEqual(gamma_y[len(gamma_y) // 2], var, places=2)
+ self.assertAlmostEqual(gamma_y[-1], var, places=2)
+
+ mask = np.zeros_like(field)
+ mask[0, 0] = 1
+ field = np.ma.masked_array(field, mask=mask)
+ gamma_x = variogram.vario_estimate_structured(field_ma, direction="x")
+ gamma_y = variogram.vario_estimate_structured(field_ma, direction="y")
+ self.assertAlmostEqual(gamma_x[0], 0.0, places=2)
+ self.assertAlmostEqual(gamma_y[0], 0.0, places=2)
def test_masked_3d(self):
rng = np.random.RandomState(1479373475)
@@ -160,45 +112,30 @@ def test_masked_3d(self):
mask = np.zeros_like(field)
field_ma = np.ma.masked_array(field, mask=mask)
- try:
- gamma_x = variogram.vario_estimate_structured(
- field_ma, direction="x"
- )
- gamma_y = variogram.vario_estimate_structured(
- field_ma, direction="y"
- )
- gamma_z = variogram.vario_estimate_structured(
- field_ma, direction="z"
- )
-
- var = 1.0 / 12.0
- self.assertAlmostEqual(gamma_x[0], 0.0, places=2)
- self.assertAlmostEqual(gamma_x[len(gamma_x) // 2], var, places=2)
- self.assertAlmostEqual(gamma_x[-1], var, places=2)
- self.assertAlmostEqual(gamma_y[0], 0.0, places=2)
- self.assertAlmostEqual(gamma_y[len(gamma_y) // 2], var, places=2)
- self.assertAlmostEqual(gamma_y[-1], var, places=2)
- self.assertAlmostEqual(gamma_z[0], 0.0, places=2)
- self.assertAlmostEqual(gamma_z[len(gamma_y) // 2], var, places=2)
- self.assertAlmostEqual(gamma_z[-1], var, places=2)
-
- mask = np.zeros_like(field)
- mask[0, 0, 0] = 1
- field = np.ma.masked_array(field, mask=mask)
- gamma_x = variogram.vario_estimate_structured(
- field_ma, direction="x"
- )
- gamma_y = variogram.vario_estimate_structured(
- field_ma, direction="y"
- )
- gamma_z = variogram.vario_estimate_structured(
- field_ma, direction="z"
- )
- self.assertAlmostEqual(gamma_x[0], 0.0, places=2)
- self.assertAlmostEqual(gamma_y[0], 0.0, places=2)
- self.assertAlmostEqual(gamma_z[0], 0.0, places=2)
- except NotImplementedError as e:
- pass
+ gamma_x = variogram.vario_estimate_structured(field_ma, direction="x")
+ gamma_y = variogram.vario_estimate_structured(field_ma, direction="y")
+ gamma_z = variogram.vario_estimate_structured(field_ma, direction="z")
+
+ var = 1.0 / 12.0
+ self.assertAlmostEqual(gamma_x[0], 0.0, places=2)
+ self.assertAlmostEqual(gamma_x[len(gamma_x) // 2], var, places=2)
+ self.assertAlmostEqual(gamma_x[-1], var, places=2)
+ self.assertAlmostEqual(gamma_y[0], 0.0, places=2)
+ self.assertAlmostEqual(gamma_y[len(gamma_y) // 2], var, places=2)
+ self.assertAlmostEqual(gamma_y[-1], var, places=2)
+ self.assertAlmostEqual(gamma_z[0], 0.0, places=2)
+ self.assertAlmostEqual(gamma_z[len(gamma_y) // 2], var, places=2)
+ self.assertAlmostEqual(gamma_z[-1], var, places=2)
+
+ mask = np.zeros_like(field)
+ mask[0, 0, 0] = 1
+ field = np.ma.masked_array(field, mask=mask)
+ gamma_x = variogram.vario_estimate_structured(field_ma, direction="x")
+ gamma_y = variogram.vario_estimate_structured(field_ma, direction="y")
+ gamma_z = variogram.vario_estimate_structured(field_ma, direction="z")
+ self.assertAlmostEqual(gamma_x[0], 0.0, places=2)
+ self.assertAlmostEqual(gamma_y[0], 0.0, places=2)
+ self.assertAlmostEqual(gamma_z[0], 0.0, places=2)
def test_uncorrelated_2d(self):
x = np.linspace(0.0, 100.0, 80)
@@ -207,19 +144,37 @@ def test_uncorrelated_2d(self):
rng = np.random.RandomState(1479373475)
field = rng.rand(len(x), len(y))
- try:
- gamma_x = variogram.vario_estimate_structured(field, direction="x")
- gamma_y = variogram.vario_estimate_structured(field, direction="y")
+ gamma_x = variogram.vario_estimate_structured(field, direction="x")
+ gamma_y = variogram.vario_estimate_structured(field, direction="y")
+
+ var = 1.0 / 12.0
+ self.assertAlmostEqual(gamma_x[0], 0.0, places=2)
+ self.assertAlmostEqual(gamma_x[len(gamma_x) // 2], var, places=2)
+ self.assertAlmostEqual(gamma_x[-1], var, places=2)
+ self.assertAlmostEqual(gamma_y[0], 0.0, places=2)
+ self.assertAlmostEqual(gamma_y[len(gamma_y) // 2], var, places=2)
+ self.assertAlmostEqual(gamma_y[-1], var, places=2)
+
+ def test_uncorrelated_cressie_2d(self):
+ x = np.linspace(0.0, 100.0, 80)
+ y = np.linspace(0.0, 100.0, 60)
+
+ rng = np.random.RandomState(1479373475)
+ field = rng.rand(len(x), len(y))
+
+ gamma_x = variogram.vario_estimate_structured(
+ field, direction="x", estimator="cressie"
+ )
+ gamma_y = variogram.vario_estimate_structured(
+ field, direction="y", estimator="cressie"
+ )
- var = 1.0 / 12.0
- self.assertAlmostEqual(gamma_x[0], 0.0, places=2)
- self.assertAlmostEqual(gamma_x[len(gamma_x) // 2], var, places=2)
- self.assertAlmostEqual(gamma_x[-1], var, places=2)
- self.assertAlmostEqual(gamma_y[0], 0.0, places=2)
- self.assertAlmostEqual(gamma_y[len(gamma_y) // 2], var, places=2)
- self.assertAlmostEqual(gamma_y[-1], var, places=2)
- except NotImplementedError as e:
- pass
+ # TODO figure out what is going on here
+ var = 0.177
+ self.assertAlmostEqual(gamma_x[0], 0.0, places=1)
+ self.assertAlmostEqual(gamma_x[len(gamma_x) // 2], var, places=1)
+ self.assertAlmostEqual(gamma_y[0], 0.0, places=1)
+ self.assertAlmostEqual(gamma_y[len(gamma_y) // 2], var, places=1)
def test_uncorrelated_3d(self):
x = np.linspace(0.0, 100.0, 30)
@@ -229,17 +184,14 @@ def test_uncorrelated_3d(self):
rng = np.random.RandomState(1479373475)
field = rng.rand(len(x), len(y), len(z))
- try:
- gamma = variogram.vario_estimate_structured(field, "x")
- gamma = variogram.vario_estimate_structured(field, "y")
- gamma = variogram.vario_estimate_structured(field, "z")
+ gamma = variogram.vario_estimate_structured(field, "x")
+ gamma = variogram.vario_estimate_structured(field, "y")
+ gamma = variogram.vario_estimate_structured(field, "z")
- var = 1.0 / 12.0
- self.assertAlmostEqual(gamma[0], 0.0, places=2)
- self.assertAlmostEqual(gamma[len(gamma) // 2], var, places=2)
- self.assertAlmostEqual(gamma[-1], var, places=2)
- except NotImplementedError as e:
- pass
+ var = 1.0 / 12.0
+ self.assertAlmostEqual(gamma[0], 0.0, places=2)
+ self.assertAlmostEqual(gamma[len(gamma) // 2], var, places=2)
+ self.assertAlmostEqual(gamma[-1], var, places=2)
def test_directions_2d(self):
x = np.linspace(0.0, 20.0, 100)
@@ -252,29 +204,18 @@ def test_directions_2d(self):
# random values repeated along x-axis
field_y = np.tile(y_rand, (len(x), 1))
- try:
- gamma_x_x = variogram.vario_estimate_structured(
- field_x, direction="x"
- )
- gamma_x_y = variogram.vario_estimate_structured(
- field_x, direction="y"
- )
-
- gamma_y_x = variogram.vario_estimate_structured(
- field_y, direction="x"
- )
- gamma_y_y = variogram.vario_estimate_structured(
- field_y, direction="y"
- )
-
- self.assertAlmostEqual(gamma_x_y[1], 0.0)
- self.assertAlmostEqual(gamma_x_y[len(gamma_x_y) // 2], 0.0)
- self.assertAlmostEqual(gamma_x_y[-1], 0.0)
- self.assertAlmostEqual(gamma_y_x[1], 0.0)
- self.assertAlmostEqual(gamma_y_x[len(gamma_x_y) // 2], 0.0)
- self.assertAlmostEqual(gamma_y_x[-1], 0.0)
- except NotImplementedError as e:
- pass
+ gamma_x_x = variogram.vario_estimate_structured(field_x, direction="x")
+ gamma_x_y = variogram.vario_estimate_structured(field_x, direction="y")
+
+ gamma_y_x = variogram.vario_estimate_structured(field_y, direction="x")
+ gamma_y_y = variogram.vario_estimate_structured(field_y, direction="y")
+
+ self.assertAlmostEqual(gamma_x_y[1], 0.0)
+ self.assertAlmostEqual(gamma_x_y[len(gamma_x_y) // 2], 0.0)
+ self.assertAlmostEqual(gamma_x_y[-1], 0.0)
+ self.assertAlmostEqual(gamma_y_x[1], 0.0)
+ self.assertAlmostEqual(gamma_y_x[len(gamma_x_y) // 2], 0.0)
+ self.assertAlmostEqual(gamma_y_x[-1], 0.0)
def test_directions_3d(self):
x = np.linspace(0.0, 10.0, 20)
@@ -289,56 +230,36 @@ def test_directions_3d(self):
field_y = np.tile(y_rand.reshape((1, len(y), 1)), (len(x), 1, len(z)))
field_z = np.tile(z_rand.reshape((1, 1, len(z))), (len(x), len(y), 1))
- try:
- gamma_x_x = variogram.vario_estimate_structured(
- field_x, direction="x"
- )
- gamma_x_y = variogram.vario_estimate_structured(
- field_x, direction="y"
- )
- gamma_x_z = variogram.vario_estimate_structured(
- field_x, direction="z"
- )
-
- gamma_y_x = variogram.vario_estimate_structured(
- field_y, direction="x"
- )
- gamma_y_y = variogram.vario_estimate_structured(
- field_y, direction="y"
- )
- gamma_y_z = variogram.vario_estimate_structured(
- field_y, direction="z"
- )
-
- gamma_z_x = variogram.vario_estimate_structured(
- field_z, direction="x"
- )
- gamma_z_y = variogram.vario_estimate_structured(
- field_z, direction="y"
- )
- gamma_z_z = variogram.vario_estimate_structured(
- field_z, direction="z"
- )
- self.assertAlmostEqual(gamma_x_y[1], 0.0)
- self.assertAlmostEqual(gamma_x_y[len(gamma_x_y) // 2], 0.0)
- self.assertAlmostEqual(gamma_x_y[-1], 0.0)
- self.assertAlmostEqual(gamma_x_z[1], 0.0)
- self.assertAlmostEqual(gamma_x_z[len(gamma_x_y) // 2], 0.0)
- self.assertAlmostEqual(gamma_x_z[-1], 0.0)
- self.assertAlmostEqual(gamma_y_x[1], 0.0)
- self.assertAlmostEqual(gamma_y_x[len(gamma_x_y) // 2], 0.0)
- self.assertAlmostEqual(gamma_y_x[-1], 0.0)
- self.assertAlmostEqual(gamma_y_z[1], 0.0)
- self.assertAlmostEqual(gamma_y_z[len(gamma_x_y) // 2], 0.0)
- self.assertAlmostEqual(gamma_y_z[-1], 0.0)
- self.assertAlmostEqual(gamma_z_x[1], 0.0)
- self.assertAlmostEqual(gamma_z_x[len(gamma_x_y) // 2], 0.0)
- self.assertAlmostEqual(gamma_z_x[-1], 0.0)
- self.assertAlmostEqual(gamma_z_y[1], 0.0)
- self.assertAlmostEqual(gamma_z_y[len(gamma_x_y) // 2], 0.0)
- self.assertAlmostEqual(gamma_z_y[-1], 0.0)
- except NotImplementedError as e:
- pass
+ gamma_x_x = variogram.vario_estimate_structured(field_x, direction="x")
+ gamma_x_y = variogram.vario_estimate_structured(field_x, direction="y")
+ gamma_x_z = variogram.vario_estimate_structured(field_x, direction="z")
+
+ gamma_y_x = variogram.vario_estimate_structured(field_y, direction="x")
+ gamma_y_y = variogram.vario_estimate_structured(field_y, direction="y")
+ gamma_y_z = variogram.vario_estimate_structured(field_y, direction="z")
+
+ gamma_z_x = variogram.vario_estimate_structured(field_z, direction="x")
+ gamma_z_y = variogram.vario_estimate_structured(field_z, direction="y")
+ gamma_z_z = variogram.vario_estimate_structured(field_z, direction="z")
+
+ self.assertAlmostEqual(gamma_x_y[1], 0.0)
+ self.assertAlmostEqual(gamma_x_y[len(gamma_x_y) // 2], 0.0)
+ self.assertAlmostEqual(gamma_x_y[-1], 0.0)
+ self.assertAlmostEqual(gamma_x_z[1], 0.0)
+ self.assertAlmostEqual(gamma_x_z[len(gamma_x_y) // 2], 0.0)
+ self.assertAlmostEqual(gamma_x_z[-1], 0.0)
+ self.assertAlmostEqual(gamma_y_x[1], 0.0)
+ self.assertAlmostEqual(gamma_y_x[len(gamma_x_y) // 2], 0.0)
+ self.assertAlmostEqual(gamma_y_x[-1], 0.0)
+ self.assertAlmostEqual(gamma_y_z[1], 0.0)
+ self.assertAlmostEqual(gamma_y_z[len(gamma_x_y) // 2], 0.0)
+ self.assertAlmostEqual(gamma_y_z[-1], 0.0)
+ self.assertAlmostEqual(gamma_z_x[1], 0.0)
+ self.assertAlmostEqual(gamma_z_x[len(gamma_x_y) // 2], 0.0)
+ self.assertAlmostEqual(gamma_z_x[-1], 0.0)
+ self.assertAlmostEqual(gamma_z_y[1], 0.0)
+ self.assertAlmostEqual(gamma_z_y[len(gamma_x_y) // 2], 0.0)
+ self.assertAlmostEqual(gamma_z_y[-1], 0.0)
def test_exceptions(self):
x = np.linspace(0.0, 10.0, 20)
diff --git a/tests/test_variogram_unstructured.py b/tests/test_variogram_unstructured.py
index 4901e462..c5a59a85 100644
--- a/tests/test_variogram_unstructured.py
+++ b/tests/test_variogram_unstructured.py
@@ -1,23 +1,12 @@
-#!/usr/bin/env python
# -*- coding: utf-8 -*-
"""
This is a unittest of the variogram module.
"""
-from __future__ import division, absolute_import, print_function
-import sys
import unittest
import numpy as np
from gstools import vario_estimate_unstructured
-PY3 = sys.version_info[0] == 3
-# in python3 "long" was replaced with "int"
-# https://docs.python.org/3.3/whatsnew/3.0.html#integers
-if PY3:
- LONGTYPE = int
-else:
- LONGTYPE = long
-
class TestVariogramUnstructured(unittest.TestCase):
def setUp(self):
@@ -40,13 +29,6 @@ def test_ints(self):
bin_centres, gamma = vario_estimate_unstructured([x], z, bins)
self.assertAlmostEqual(gamma[0], 50.0, places=4)
- def test_longs(self):
- x = np.arange(1, 5, 1, dtype=LONGTYPE)
- z = np.array((10, 20, 30, 40), dtype=LONGTYPE)
- bins = np.arange(1, 11, 1, dtype=LONGTYPE)
- bin_centres, gamma = vario_estimate_unstructured([x], z, bins)
- self.assertAlmostEqual(gamma[0], 50.0, places=4)
-
def test_np_int(self):
x = np.arange(1, 5, 1, dtype=np.int)
z = np.array((10, 20, 30, 40), dtype=np.int)
@@ -65,13 +47,13 @@ def test_mixed(self):
self.assertAlmostEqual(gamma[0], 0.4917, places=4)
x = np.arange(1, 5, 1, dtype=np.double)
- z = np.array((10, 20, 30, 40), dtype=LONGTYPE)
+ z = np.array((10, 20, 30, 40), dtype=int)
bins = np.arange(1, 11, 1, dtype=int)
bin_centres, gamma = vario_estimate_unstructured([x], z, bins)
self.assertAlmostEqual(gamma[0], 50.0, places=4)
x = np.arange(1, 5, 1, dtype=np.double)
- z = np.array((10, 20, 30, 40), dtype=LONGTYPE)
+ z = np.array((10, 20, 30, 40), dtype=int)
bins = np.arange(1, 11, 1, dtype=np.double)
bin_centres, gamma = vario_estimate_unstructured([x], z, bins)
self.assertAlmostEqual(gamma[0], 50.0, places=4)
 +
+ +
+