| title | author |
|---|---|
Part I: Introduction |
Laurent Gatto |
- Coding style(s)
- Interactive use and programming
- Environments
- Tidy data
- Computing on the language
Computers are cheap, and thinking hurts. -- Uwe Ligges
Simplicity, readability and consistency are a long way towards robust code.
Why?
Good coding style is like using correct punctuation. You can manage without it, but it sure makes things easier to read. -- Hadley Wickham
for consistency and readability.
- Place spaces around all infix operators (
=,+,-,<-, etc., but not:) and after a comma (x[i, j]). - Spaces before
(and after); not for function. - Use
<-rather than=. - Limit your code to 80 characters per line
- Indentation: do not use tabs, use 2 (HW)/4 (Bioc) spaces
- Function names: use verbs
- Variable names: camelCaps (Bioc)/
_(HW) (but not a.) - Prefix non-exported functions with a ‘.’ (Bioc).
- Class names: start with a capital
- Comments:
#or##(from emacs)
library("formatR")
tidy_source(text = "a=1+1;a # print the value
matrix ( rnorm(10),5)",
arrow = TRUE)## a <- 1 + 1
## a # print the value
## matrix(rnorm(10), 5)
$ R CMD BiocCheck package_1.0.0.tgz
* Checking function lengths................
The longest function is 677 lines long
The longest 5 functions are:
* Checking formatting of DESCRIPTION, NAMESPACE, man pages, R source,
and vignette source...
* CONSIDER: Shortening lines; 616 lines (11%) are > 80 characters
long.
* CONSIDER: Replacing tabs with 4 spaces; 3295 lines (60%) contain
tabs.
* CONSIDER: Indenting lines with a multiple of 4 spaces; 162 lines
(2%) are not.
Moving from using R to programming R is abstraction, automation, generalisation.
head(cars)
head(cars[, 1])
head(cars[, 1, drop = FALSE])df1 <- data.frame(x = 1:3, y = LETTERS[1:3])
sapply(df1, class)
df2 <- data.frame(x = 1:3, y = Sys.time() + 1:3)
sapply(df2, class)Rather use a form where the return data structure is known...
lapply(df1, class)
lapply(df2, class)or that will break if the result is not what is exected
vapply(df1, class, "1")
vapply(df2, class, "1")- pass-by-value copy-on-modify
- pass-by-reference: environments, S4 Reference Classes
x <- 1
f <- function(x) {
x <- 2
x
}
x
f(x)
x- Data structure that enables scoping (see later).
- Have reference semantics
- Useful data structure on their own
An environment associates, or binds, names to values in memory. Variables in an environment are hence called bindings.
e <- new.env()
e$a <- 1
e$b <- LETTERS[1:5]
e$c <- TRUE
e$d <- meane$a <- e$b
e$a <- LETTERS[1:5]- Objects in environments have unique names
- Objects in different environments can of course have identical names
- Objects in an environment have no order
- Environments have parents
An environment is composed of a frame that contains the name-object bindings and a parent (enclosing) environment.
Every environment has a parent (enclosing) environment
e <- new.env()
parent.env(e)Current environment
environment()
parent.env(globalenv())
parent.env(parent.env(globalenv()))Noteworthy environments
globalenv()
emptyenv()
baseenv()All parent of R_GlobalEnv:
search()
as.environment("package:stats")Listing objects in an environment
ls() ## default is R_GlobalEnv
ls(envir = e)
ls(pos = 1)search()Note: Every time a package is loaded with library, it is inserted in
the search path after the R_GlobalEnv.
- In addition to
$, one can also use[[,getandassign. - To check if a name exists in an environmet (or in any or its
parents), one can use
exists. - Compare two environments with
identical(not==).
Question Are e1 and e2 below identical?
e1 <- new.env()
e2 <- new.env()
e1$a <- 1:10
e2$a <- e1$aWhat about e1 and e3?
e3 <- e1
e3
e1
identical(e1, e3)e <- new.env()
e$a <- 1
e$b <- 2 ## add
e$a <- 10 ## modifyLocking an environment stops from adding new bindings:
lockEnvironment(e)
e$k <- 1
e$a <- 100Locking bindings stops from modifying bindings with en envionment:
lockBinding("a", e)
e$a <- 10
e$b <- 10
lockEnvironment(e, bindings = TRUE)
e$b <- 1Reproduce the following environments and variables in R.
pryr::where() implements the regular scoping rules to find in which
environment a binding is defined.
e <- new.env()
e$foo <- 1
bar <- 2
where("foo")
where("bar")
where("foo", env = e)
where("bar", env = e)[Lexical comes from lexical analysis in computer science, which is the conversion of characters (code) into a sequence of meaningful (for the computer) tokens.]
Definition: Rules that define how R looks up values for a given name/symbol.
- Objects in environments have unique names
- Objects in different environments can of course have identical names.
- If a name is not found in the current environment, it is looked up in the parent (enclosing) from.
- If it is not found in the parent (enclosing) frame, it is looked up in the parent's parent frame, and so on...
search()
mean <- function(x) cat("The mean is", sum(x)/length(x), "\n")
mean(1:10)
base::mean(1:10)
rm(mean)
mean(1:10)-
<-assigns/creates in the current environment -
<<-(deep assignment) never creates/updates a variable in the current environment, but modifies an existing variable in the current or first enclosing environment where that name is defined. -
If
<<-does not find the name, it will create the variable in the global environment.
library("fortunes")
fortune(174)##
## I wish <<- had never been invented, as it makes an esoteric and dangerous
## feature of the language *seem* normal and reasonable. If you want to dumb
## down R/S into a macro language, this is the operator for you.
## -- Bill Venables
## R-help (July 2001)
rm(list = ls())
x
f1 <- function() x <<- 1
f1()
xf2 <- function() x <<- 2
f2()
xf3 <- function() x <- 10
f3()
xf4 <- function(x) x <-10
f4(x)
xMost environments are created when creating and calling functions. They are also used in packages as package and namespace environments.
There are several reasons to create then manually.
- Reference semantics
- Avoiding copies
- Package state
- As a hashmap for fast name lookup
modify <- function(x) {
x$a <- 2
invisible(TRUE)
}x_l <- list(a = 1)
modify(x_l)
x_l$ax_e <- new.env()
x_e$a <- 1
modify(x_e)
x_e$aTip: when setting up environments, it is advised to set to parent
(enclosing) environment to be emptyenv(), to avoid accidentally
inheriting objects from somewhere else on the search path.
e <- new.env()
e$a <- 1
e
parent.env(e)
parent.env(e) <- emptyenv()
parent.env(e)
eor directly
e <- new.env(parent.env = empty.env())What is going to happen when we access "x" in the four cases below?
x <- 1
e1 <- new.env()
get("x", envir = e1)get("x", envir = e1, inherits = FALSE)e2 <- new.env(parent = emptyenv())
get("x", envir = e2)get("x", envir = e1, inherits = FALSE)Since environments have reference semantics, they are not copied. When passing an environment as function argument (directly, or as part of a more complex data structure), it is not copied: all its values are accessible within the function and can be persistently modified.
e <- new.env()
e$x <- 1
f <- function(myenv) myenv$x <- 2
f(e)
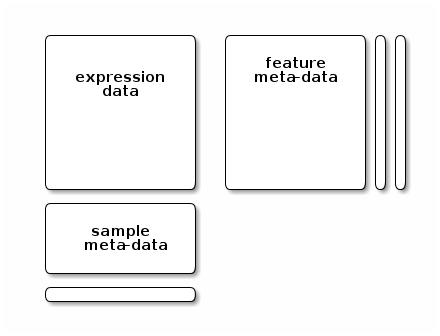
e$xThis is used in the eSet class family to store the expression data.
library("Biobase")
getClass("eSet")
getClass("AssayData")
new("ExpressionSet")Explicit envirionments are also useful to preserve state or define constants-like variables in a package. One can then set getters and setters for users to access the variables within that private envionment.
Colour management in pRoloc:
.pRolocEnv <- new.env(parent=emptyenv(), hash=TRUE)
stockcol <- c("#E41A1C", "#377EB8", "#238B45", "#FF7F00", "#FFD700", "#333333",
"#00CED1", "#A65628", "#F781BF", "#984EA3", "#9ACD32", "#B0C4DE",
"#00008A", "#8B795E", "#FDAE6B", "#66C2A5", "#276419", "#CD8C95",
"#6A51A3", "#EEAD0E", "#0000FF", "#9ACD32", "#CD6090", "#CD5B45",
"#8E0152", "#808000", "#67000D", "#3F007D", "#6BAED6", "#FC9272")
assign("stockcol", stockcol, envir = .pRolocEnv)
getStockcol <- function() get("stockcol", envir = .pRolocEnv)
setStockcol <- function(cols) {
if (is.null(cols)) {
assign("stockcol", stockcol, envir = .pRolocEnv)
} else {
assign("stockcol", cols, envir = .pRolocEnv)
}
}and in plotting functions (we will see the missing in more details later):
...
if (missing(col))
col <- getStockcol()
...Hadley's tip: Invisibly returning the old value from
setStockcol <- function(cols) {
prevcols <- getStockcol()
if (is.null(cols)) {
assign("stockcol", stockcol, envir = .pRolocEnv)
} else {
assign("stockcol", cols, envir = .pRolocEnv)
}
invisible(prevcols)
}Hadley Wickham, Tidy Data, Vol. 59, Issue 10, Sep 2014, Journal of Statistical Software. http://www.jstatsoft.org/v59/i10.
Tidy datasets are easy to manipulate, model and visualize, and have a specific structure: each variable is a column, each observation is a row, and each type of observational unit is a table.
Tidy data also makes it easier to develop tidy tools for data analysis, tools that both input and output tidy datasets.
dply::selectselect columnsdlpy::filterselect rowsdplyr:mutatecreate new columnsdpplyr:group_bysplit-apply-combinedlpyr:summarisecollapse each group into a single-row summary of that groupmagrittr::%>%piping
library("dplyr")
surveys <- read.csv("http://datacarpentry.github.io/dc_zurich/data/portal_data_joined.csv")
head(surveys)
surveys %>%
filter(weight < 5) %>%
select(species_id, sex, weight)
surveys %>%
mutate(weight_kg = weight / 1000) %>%
filter(!is.na(weight)) %>%
head
surveys %>%
group_by(sex) %>%
tally()
surveys %>%
group_by(sex, species_id) %>%
summarize(mean_weight = mean(weight, na.rm = TRUE))
surveys %>%
group_by(sex, species_id) %>%
summarize(mean_weight = mean(weight, na.rm = TRUE),
min_weight = min(weight, na.rm = TRUE)) %>%
filter(!is.nan(mean_weight))Hadley Wickham (@hadleywickham) tweeted at 8:45 pm on Fri, Feb 12, 2016: @mark_scheuerell @drob the importance of tidy data is not the specific form, but the consistency (https://twitter.com/hadleywickham/status/698246671629549568?s=09)
- Well-formatted and well-documented
S4class S4as input -(function)->S4as output
Quote an expression, don't evaluate it:
quote(1:10)
quote(paste(letters, LETTERS, sep = "-"))Evaluate an expression in a specific environment:
eval(quote(1 + 1))
eval(quote(1:10))
x <- 10
eval(quote(x + 1))
e <- new.env()
e$x <- 1
eval(quote(x + 1), env = e)
eval(quote(x), list(x = 30))
dfr <- data.frame(x = 1:10, y = LETTERS[1:10])
eval(quote(sum(x)), dfr)Substitute any variables bound in env, but don't evaluate the
expression:
x <- 10
substitute(sqrt(x))
e <- new.env()
e$x <- 1
substitute(sqrt(x), env = e)Parse, but don't evaluate an expression:
parse(text = "1:10")
parse(file = "lineprof-example.R")Turn an unevaluated expressions into character strings:
x <- 123
deparse(substitute(x))foo <- "bar"
as.name(foo)
string <- "1:10"
parse(text=string)
eval(parse(text=string))And with assign and get
varName1 <- "varName2"
assign(varName1, "123")
varName1
get(varName1)
varName2Using substitute and deparse
test <- function(x) {
y <- deparse(substitute(x))
print(y)
print(x)
}
var <- c("one","two","three")
test(var)